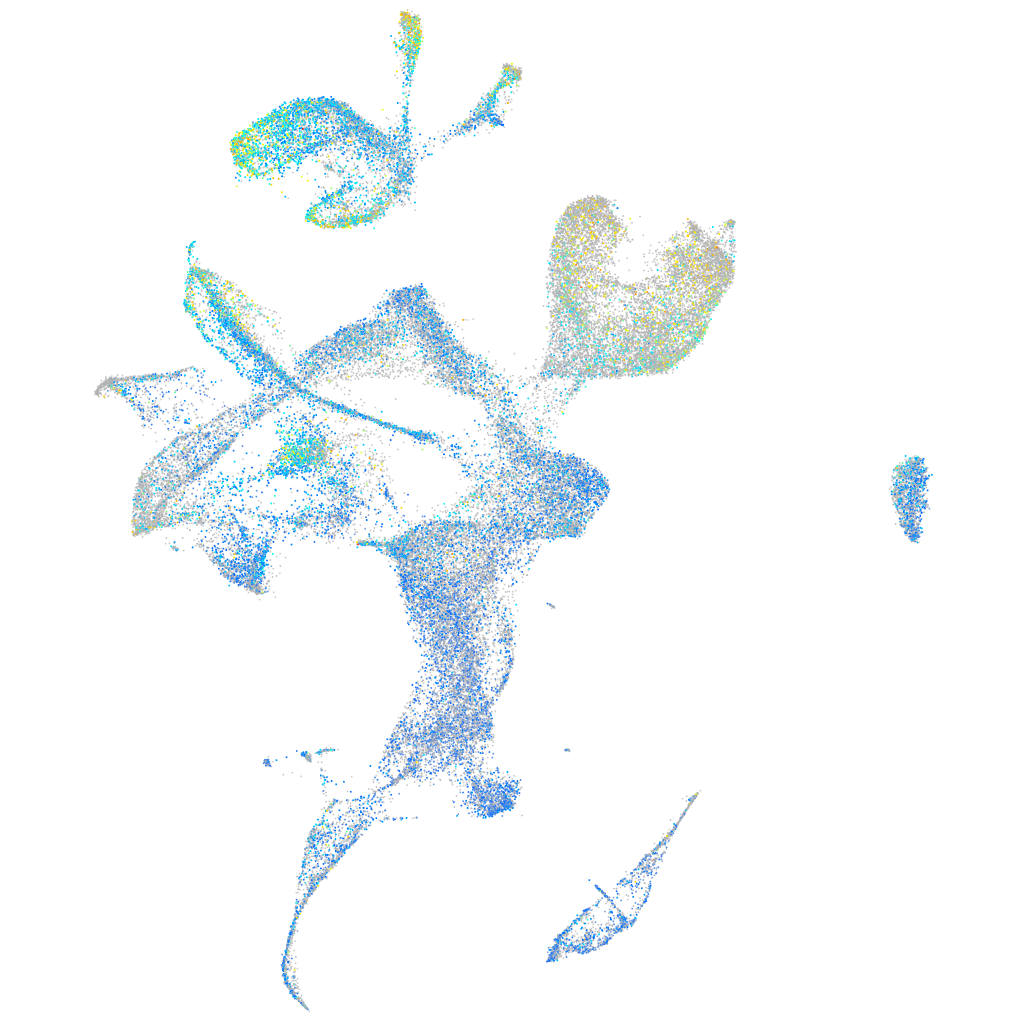FUN14 domain containing 1
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| stxbp1a | 0.312 | hmgb2a | -0.169 |
| ywhag2 | 0.307 | pcna | -0.100 |
| syt1a | 0.291 | ccnd1 | -0.095 |
| stmn1b | 0.272 | rrm1 | -0.095 |
| stx1b | 0.271 | zgc:165555.3 | -0.092 |
| slc32a1 | 0.271 | dut | -0.092 |
| gng3 | 0.271 | stmn1a | -0.092 |
| sncb | 0.259 | syt5b | -0.091 |
| elavl3 | 0.258 | crx | -0.089 |
| eno2 | 0.258 | chaf1a | -0.088 |
| si:ch73-119p20.1 | 0.255 | hist1h4l | -0.088 |
| zgc:65894 | 0.255 | rrm2 | -0.086 |
| rbfox1 | 0.254 | si:dkey-261m9.17 | -0.086 |
| rtn1b | 0.252 | mcm7 | -0.085 |
| sv2a | 0.251 | CABZ01005379.1 | -0.084 |
| snap25a | 0.248 | mki67 | -0.084 |
| slc6a1a | 0.248 | msi1 | -0.083 |
| sprn | 0.242 | otx5 | -0.083 |
| zgc:153426 | 0.236 | neurod4 | -0.083 |
| syn2a | 0.234 | mibp | -0.082 |
| mir181b-3 | 0.233 | mdka | -0.082 |
| slc35g2b | 0.231 | dek | -0.082 |
| stmn2a | 0.226 | lig1 | -0.081 |
| gabrb4 | 0.226 | nutf2l | -0.081 |
| dtnbp1b | 0.218 | zgc:110425 | -0.081 |
| slc6a1b | 0.218 | fabp7a | -0.081 |
| gng2 | 0.214 | lbr | -0.080 |
| celf5a | 0.209 | ccna2 | -0.079 |
| ppp1r14ba | 0.208 | her15.1 | -0.079 |
| carmil2 | 0.207 | ahcy | -0.079 |
| atp1b3a | 0.207 | zgc:110216 | -0.078 |
| nrxn1a | 0.207 | esco2 | -0.078 |
| st8sia5 | 0.207 | rps29 | -0.077 |
| tfap2a | 0.206 | zgc:110540 | -0.077 |
| calm2b | 0.205 | tuba8l | -0.077 |