forkhead box F2b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 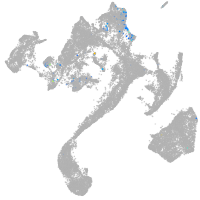 |
mural cells / non-skeletal muscle  |
mesenchyme 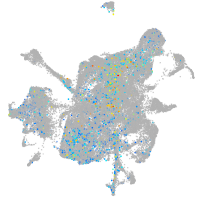 |
hematopoietic / vasculature 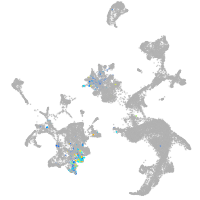 |
pronephros 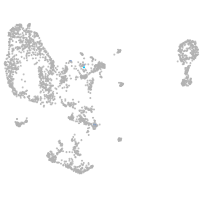 |
neural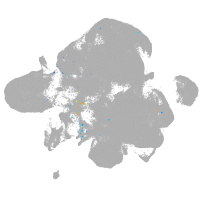 |
spinal cord / glia |
eye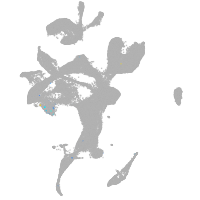 |
taste / olfactory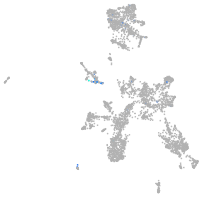 |
otic / lateral line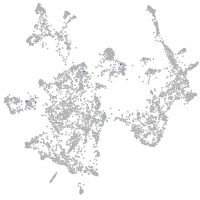 |
epidermis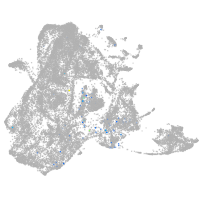 |
periderm |
fin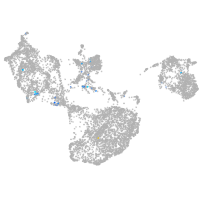 |
ionocytes / mucous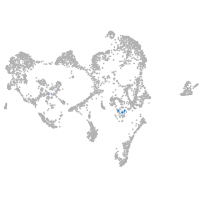 |
pigment cells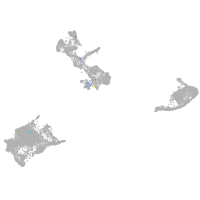 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nptx2a | 0.443 | paics | -0.031 |
| rasl12 | 0.409 | syngr1a | -0.028 |
| plpp2a | 0.384 | gpr143 | -0.028 |
| tbx18 | 0.336 | tspan36 | -0.026 |
| abcc9 | 0.325 | gart | -0.025 |
| si:ch211-202p1.5 | 0.316 | agtrap | -0.024 |
| gpc5c | 0.304 | pvalb1 | -0.024 |
| ccdc3b | 0.288 | rpl11 | -0.024 |
| ehd2a | 0.281 | smim29 | -0.022 |
| ucmaa | 0.274 | C12orf75 | -0.022 |
| stac | 0.265 | rabl6b | -0.022 |
| fgfr1bl | 0.259 | gstp1 | -0.022 |
| foxf2a | 0.253 | rps8a | -0.022 |
| CU467646.6 | 0.252 | akr1b1 | -0.022 |
| LOC103909118 | 0.239 | oacyl | -0.022 |
| XLOC-043092 | 0.233 | bace2 | -0.022 |
| LOC100004160 | 0.232 | rnaseka | -0.021 |
| bmp16 | 0.227 | XLOC-019062 | -0.021 |
| LRRC75A | 0.226 | actb2 | -0.021 |
| gucy1a1 | 0.226 | tspan10 | -0.021 |
| AL935126.2 | 0.225 | pfn1 | -0.021 |
| c1qtnf6a | 0.221 | coro1ca | -0.021 |
| colec12 | 0.219 | prtfdc1 | -0.021 |
| map3k9 | 0.208 | zgc:110591 | -0.020 |
| foxf1 | 0.207 | slc2a15a | -0.020 |
| jade2 | 0.206 | si:dkey-21a6.5 | -0.020 |
| opn1mw2 | 0.205 | sptlc2a | -0.020 |
| LOC108180013 | 0.204 | rps13 | -0.020 |
| si:ch211-110p13.9 | 0.204 | slc2a11b | -0.020 |
| zgc:114118 | 0.202 | pts | -0.020 |
| bgnb | 0.193 | impdh1b | -0.019 |
| LOC101885894 | 0.192 | rab32a | -0.019 |
| fzd8a | 0.191 | nmt1a | -0.019 |
| shroom3 | 0.189 | rab38 | -0.019 |
| gdnfa | 0.187 | rps26l | -0.019 |