family with sequence similarity 117 member Ba
ZFIN 
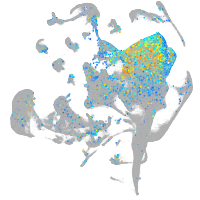
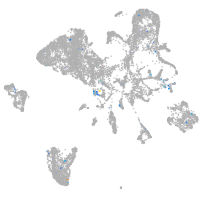
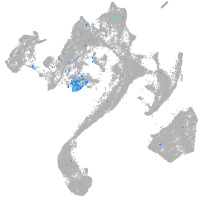
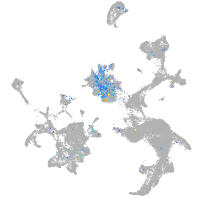
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gng3 | 0.327 | hmgb2a | -0.302 |
| sncb | 0.326 | id1 | -0.251 |
| elavl4 | 0.326 | pcna | -0.248 |
| stmn1b | 0.315 | hmga1a | -0.231 |
| stmn2a | 0.312 | hmgb2b | -0.228 |
| stx1b | 0.307 | si:dkey-151g10.6 | -0.226 |
| ywhag2 | 0.304 | stmn1a | -0.226 |
| gpm6ab | 0.303 | ranbp1 | -0.214 |
| tuba1c | 0.303 | mdka | -0.214 |
| rtn1b | 0.300 | anp32b | -0.214 |
| vamp2 | 0.293 | npm1a | -0.211 |
| rtn1a | 0.292 | XLOC-003690 | -0.208 |
| gap43 | 0.290 | selenoh | -0.207 |
| ckbb | 0.288 | chaf1a | -0.207 |
| stxbp1a | 0.288 | nop58 | -0.207 |
| zgc:65894 | 0.288 | sox3 | -0.207 |
| snap25a | 0.288 | msna | -0.206 |
| atp6v0cb | 0.286 | rplp1 | -0.206 |
| ywhah | 0.281 | ccnd1 | -0.205 |
| calm1a | 0.278 | sox19a | -0.202 |
| mllt11 | 0.274 | snu13b | -0.201 |
| atp6v1e1b | 0.272 | si:ch73-281n10.2 | -0.201 |
| tmsb2 | 0.269 | banf1 | -0.200 |
| elavl3 | 0.268 | eef1a1l1 | -0.197 |
| atp6v1g1 | 0.266 | rrm1 | -0.196 |
| gpm6aa | 0.265 | rplp2l | -0.196 |
| gnao1a | 0.261 | nop56 | -0.195 |
| gapdhs | 0.259 | mcm6 | -0.194 |
| map1aa | 0.258 | dkc1 | -0.194 |
| ptmaa | 0.256 | ran | -0.194 |
| si:dkeyp-75h12.5 | 0.256 | tuba8l | -0.194 |
| kdm6bb | 0.254 | lig1 | -0.194 |
| tuba2 | 0.254 | mcm7 | -0.193 |
| rnasekb | 0.254 | tuba8l4 | -0.193 |
| zc4h2 | 0.253 | dut | -0.192 |