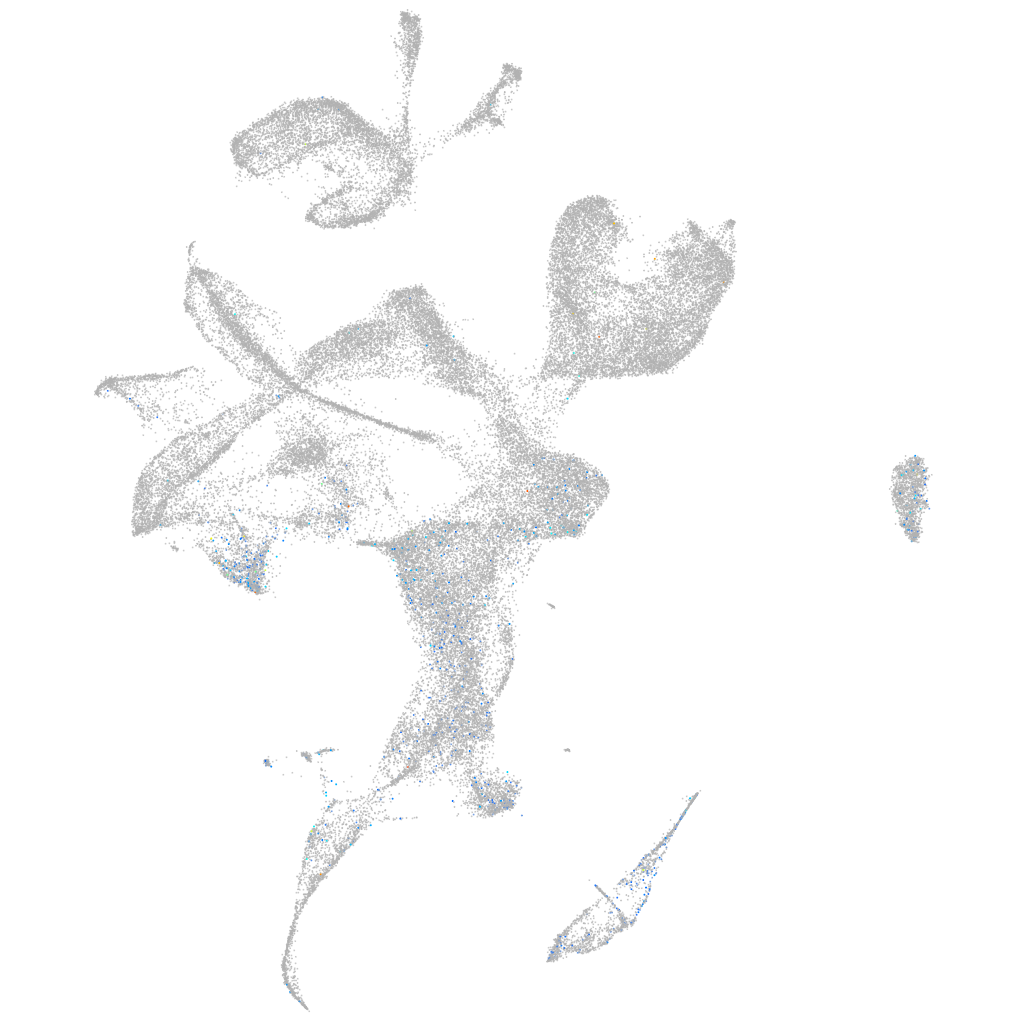
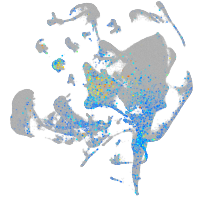
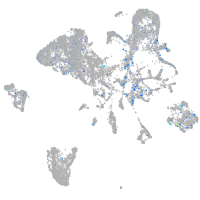
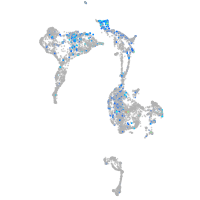
eph receptor B4b
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkeyp-73g8.5 | 0.141 | ckbb | -0.056 |
| si:dkey-269p2.1 | 0.100 | ptmab | -0.055 |
| pmp22a | 0.097 | atp6v0cb | -0.051 |
| fmoda | 0.094 | snap25b | -0.050 |
| col5a1 | 0.092 | hmgn6 | -0.048 |
| id1 | 0.089 | rnasekb | -0.047 |
| mfap2 | 0.085 | ndrg4 | -0.045 |
| pdgfra | 0.084 | gpm6ab | -0.043 |
| tpm4a | 0.084 | ptmaa | -0.042 |
| col11a1b | 0.081 | atp6v1e1b | -0.042 |
| XLOC-004932 | 0.080 | gpm6aa | -0.041 |
| ednraa | 0.079 | atp1a3a | -0.040 |
| emp2 | 0.079 | pcbp3 | -0.040 |
| colec12 | 0.077 | atp1b2a | -0.040 |
| fstl1b | 0.077 | epb41a | -0.040 |
| plcd4a | 0.077 | ppp1r14c | -0.039 |
| tmem176 | 0.077 | atpv0e2 | -0.039 |
| fgfr2 | 0.077 | syt5b | -0.039 |
| plod1a | 0.076 | calb2b | -0.039 |
| lim2.1 | 0.075 | gng13b | -0.038 |
| gja8b | 0.075 | gapdhs | -0.038 |
| aoc2 | 0.074 | atp6v1g1 | -0.038 |
| fam198a | 0.074 | gng3 | -0.037 |
| boc | 0.074 | mpp6b | -0.037 |
| rbm24a | 0.073 | otx5 | -0.037 |
| krt8 | 0.073 | vamp2 | -0.036 |
| mmp14a | 0.072 | pvalb2 | -0.036 |
| fzd2 | 0.071 | sncb | -0.036 |
| cx23 | 0.071 | pvalb1 | -0.035 |
| col6a2 | 0.071 | crx | -0.035 |
| her6 | 0.069 | olfm1b | -0.035 |
| pdgfrl | 0.069 | stmn1b | -0.034 |
| cryba1l1 | 0.069 | isl1 | -0.034 |
| thbs2a | 0.069 | scrt2 | -0.034 |
| krt18a.1 | 0.069 | tuba1c | -0.034 |