dynein regulatory complex subunit 3
ZFIN 
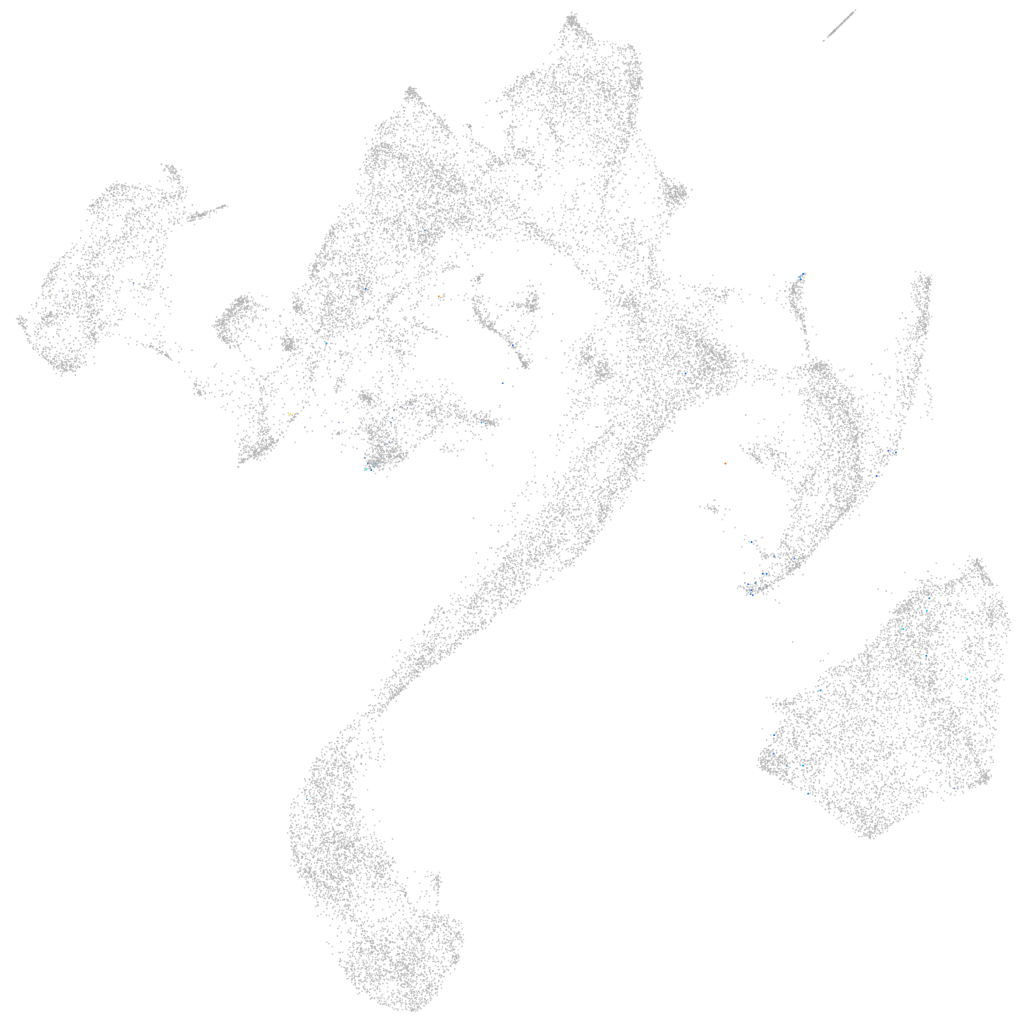
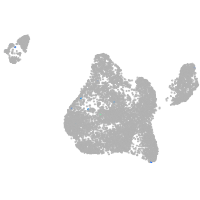
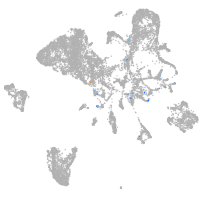
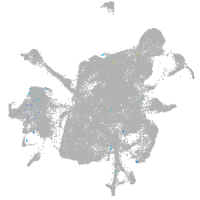
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| znf1085 | 0.289 | uba52 | -0.028 |
| wdr78 | 0.272 | rpl32 | -0.028 |
| smkr1 | 0.260 | rps28 | -0.025 |
| cfap157 | 0.244 | COX3 | -0.020 |
| spef2 | 0.243 | rpl6 | -0.020 |
| ccdc65 | 0.227 | ahnak | -0.020 |
| ccdc169 | 0.218 | rpl18 | -0.019 |
| lrrc74b | 0.208 | meox1 | -0.019 |
| glis3 | 0.207 | rps27.2 | -0.019 |
| fabp7b | 0.184 | FQ323156.1 | -0.019 |
| ak7a | 0.176 | gamt | -0.019 |
| si:dkey-223p19.2 | 0.171 | vwde | -0.018 |
| CU929150.2 | 0.164 | mt-co2 | -0.018 |
| si:dkey-27p23.3 | 0.159 | rpl5a | -0.018 |
| nme5 | 0.158 | eif5a | -0.018 |
| tbata | 0.150 | emp2 | -0.018 |
| cbln2a | 0.150 | rpl27a | -0.017 |
| bcan | 0.146 | col1a2 | -0.017 |
| dydc2 | 0.146 | rpl21 | -0.017 |
| CABZ01084347.1 | 0.141 | mt-co1 | -0.017 |
| zgc:195023 | 0.140 | slc25a3b | -0.017 |
| CR812470.2 | 0.140 | zgc:92429 | -0.017 |
| catip | 0.138 | gatm | -0.017 |
| CR848749.2 | 0.136 | gapdh | -0.017 |
| enkur | 0.134 | anxa11a | -0.016 |
| spag8 | 0.130 | rpl36a | -0.016 |
| LOC101885096 | 0.129 | CABZ01078594.1 | -0.016 |
| daw1 | 0.128 | eef1da | -0.016 |
| dnali1 | 0.127 | rps15 | -0.016 |
| spata17 | 0.127 | sparc | -0.016 |
| spata4 | 0.116 | COX5B | -0.016 |
| LOC110438143 | 0.115 | meis1a | -0.016 |
| BX530034.1 | 0.115 | cfl2 | -0.016 |
| nt5c1ba | 0.112 | mfap2 | -0.016 |
| si:dkey-246j6.3 | 0.109 | smco4 | -0.016 |