"dynein, axonemal, light chain 4a"
ZFIN 
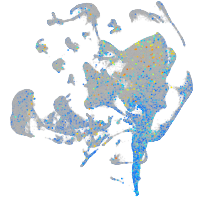
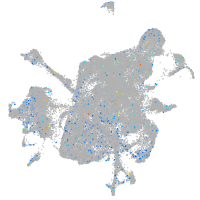
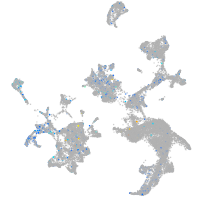
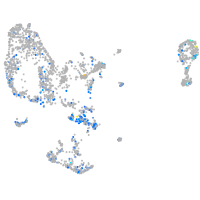
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccdc173 | 0.238 | dab2 | -0.143 |
| foxj1a | 0.231 | aldob | -0.139 |
| gas8 | 0.224 | gapdh | -0.131 |
| ribc2 | 0.218 | gcshb | -0.128 |
| XLOC-016165 | 0.217 | fbp1b | -0.127 |
| ccdc151 | 0.216 | si:dkey-33i11.9 | -0.124 |
| ttc26 | 0.214 | si:ch211-139a5.9 | -0.123 |
| BX323016.1 | 0.210 | zgc:136493 | -0.120 |
| catip | 0.208 | eno3 | -0.119 |
| zgc:158640 | 0.208 | epdl2 | -0.119 |
| cfap20 | 0.205 | grhprb | -0.118 |
| spaca9 | 0.205 | sord | -0.118 |
| dnai1.2 | 0.204 | si:dkey-28n18.9 | -0.118 |
| arhgap22 | 0.202 | sod1 | -0.118 |
| cluap1 | 0.202 | slc20a1a | -0.117 |
| morn2 | 0.201 | pnp6 | -0.116 |
| rfx2 | 0.197 | nit2 | -0.115 |
| ift140 | 0.197 | alpl | -0.115 |
| si:ch211-163l21.7 | 0.197 | suclg2 | -0.113 |
| si:dkey-114c15.5 | 0.195 | glud1b | -0.113 |
| ttc25 | 0.195 | nipsnap3a | -0.113 |
| gmnc | 0.194 | gamt | -0.112 |
| tekt2 | 0.193 | gstr | -0.110 |
| si:ch211-194m7.3 | 0.193 | dpys | -0.110 |
| spa17 | 0.192 | hao1 | -0.110 |
| CU929160.1 | 0.191 | nrip2 | -0.110 |
| ncs1a | 0.191 | upb1 | -0.109 |
| cfap45 | 0.191 | cx32.3 | -0.109 |
| ift27 | 0.191 | ASS1 | -0.109 |
| sall1b | 0.190 | cubn | -0.108 |
| dpcd | 0.189 | akr7a3 | -0.108 |
| si:ch211-248e11.2 | 0.186 | gstt1a | -0.108 |
| irak1bp1 | 0.185 | lrp2a | -0.107 |
| cetn4 | 0.184 | agxta | -0.106 |
| efcab2 | 0.183 | gpt2l | -0.106 |