"dynein, axonemal, light chain 4a"
ZFIN 
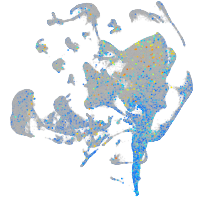
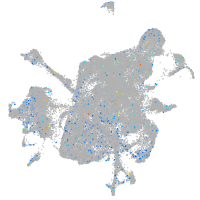
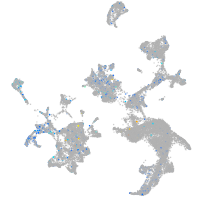
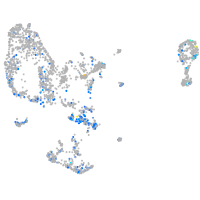
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| daw1 | 0.079 | tmsb4x | -0.044 |
| cdx4 | 0.077 | fabp3 | -0.041 |
| si:dkeyp-110a12.4 | 0.073 | ckbb | -0.035 |
| spag6 | 0.073 | LOC798783 | -0.030 |
| meig1 | 0.068 | tmsb | -0.029 |
| pou5f3 | 0.068 | nhlh2 | -0.028 |
| dpcd | 0.068 | mdkb | -0.028 |
| foxj1a | 0.067 | fabp7a | -0.028 |
| si:ch211-163l21.7 | 0.067 | CU467822.1 | -0.027 |
| smkr1 | 0.066 | elavl3 | -0.027 |
| cfap45 | 0.064 | gadd45gb.1 | -0.027 |
| dnaaf4 | 0.064 | si:ch211-57n23.4 | -0.027 |
| si:ch73-81k8.2 | 0.064 | rtn1a | -0.026 |
| cfap126 | 0.063 | CU634008.1 | -0.026 |
| hspb1 | 0.062 | marcksl1b | -0.026 |
| ccdc173 | 0.061 | dlb | -0.026 |
| ankrd45 | 0.061 | si:ch73-21g5.7 | -0.026 |
| ribc2 | 0.061 | actc1b | -0.026 |
| catip | 0.061 | scrt1a | -0.025 |
| tppp3 | 0.060 | pvalb1 | -0.025 |
| BX664625.2 | 0.060 | pvalb2 | -0.025 |
| si:ch211-155m12.5 | 0.059 | pou3f1 | -0.025 |
| LOC103910812 | 0.059 | pdzd11 | -0.024 |
| gdf6b | 0.059 | BX530077.1 | -0.023 |
| enkur | 0.059 | sox11b | -0.023 |
| spata4 | 0.059 | scrt2 | -0.023 |
| rab36 | 0.059 | myt1b | -0.023 |
| zgc:55461 | 0.059 | actb1 | -0.023 |
| rsph9 | 0.058 | thsd7aa | -0.022 |
| efcab1 | 0.058 | ascl1b | -0.022 |
| cfap298 | 0.058 | gdf11 | -0.022 |
| chrd | 0.058 | efna1b | -0.022 |
| morn3 | 0.058 | slc1a2b | -0.022 |
| cfap52 | 0.057 | si:ch211-133n4.4 | -0.022 |
| si:dkey-114c15.5 | 0.057 | hbbe1.2 | -0.021 |