doublesex and mab-3 related transcription factor 2a
ZFIN 
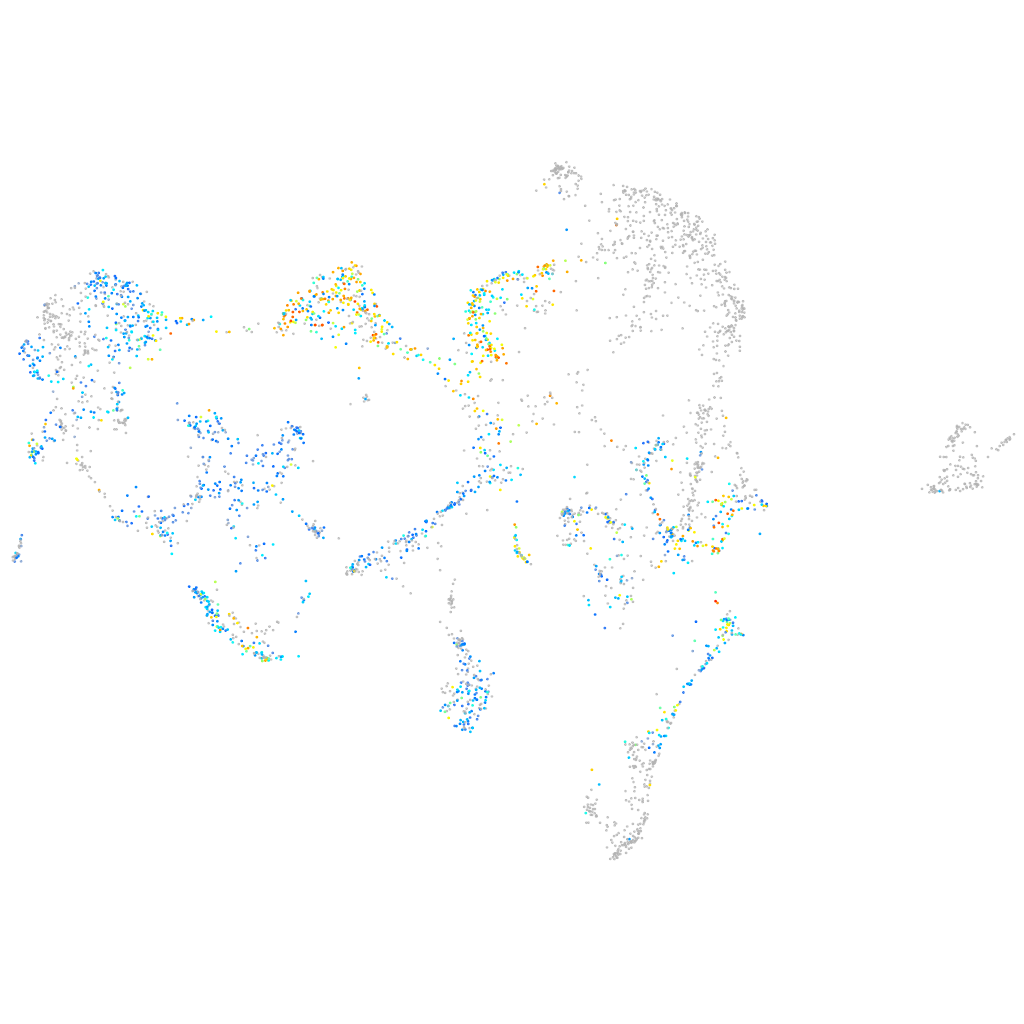
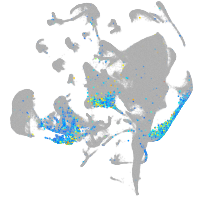
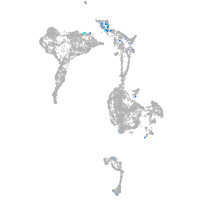
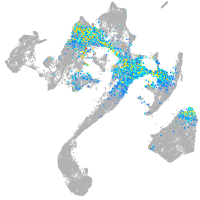
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| foxi3b | 0.572 | agr2 | -0.423 |
| ndrg3a | 0.558 | muc5.1 | -0.375 |
| si:dkey-33i11.4 | 0.555 | si:dkey-65b12.6 | -0.373 |
| atp1b1b | 0.552 | zgc:92380 | -0.339 |
| ndrg1a | 0.480 | b3gnt7 | -0.337 |
| tmx2b | 0.465 | atp2a3 | -0.331 |
| atp1a1a.2 | 0.448 | stard10 | -0.320 |
| si:ch73-31d8.2 | 0.428 | p2rx1 | -0.318 |
| gadd45ab | 0.421 | nansb | -0.315 |
| zgc:194246 | 0.417 | foxa3 | -0.303 |
| mdh1aa | 0.415 | fxyd1 | -0.302 |
| elovl1b | 0.394 | LOC110438378 | -0.295 |
| cox5aa | 0.387 | sytl2b | -0.286 |
| rgs5b | 0.377 | ucp2 | -0.283 |
| mdh2 | 0.377 | rpz5 | -0.282 |
| atp5mc3b | 0.377 | xbp1 | -0.282 |
| cox7a2a | 0.374 | muc5.2 | -0.280 |
| mat2al | 0.374 | SPDEF | -0.277 |
| LOC799574 | 0.373 | pdia3 | -0.276 |
| COX5B | 0.371 | galnt7 | -0.268 |
| si:ch211-270g19.5 | 0.370 | si:dkey-151g10.6 | -0.266 |
| sgk2a | 0.370 | mir375-2 | -0.263 |
| trim35-12 | 0.369 | cebpd | -0.262 |
| cox7b | 0.367 | gng13b | -0.261 |
| ncln | 0.367 | zgc:112994 | -0.257 |
| ddt | 0.366 | krt5 | -0.257 |
| vdac2 | 0.364 | zgc:153675 | -0.257 |
| atp5l | 0.364 | kdelr3 | -0.257 |
| prdx5 | 0.363 | crip1 | -0.252 |
| atp5f1b | 0.361 | klf3 | -0.252 |
| atp5fa1 | 0.359 | tent5bb | -0.251 |
| cox6a1 | 0.353 | anxa5b | -0.250 |
| selenow2b | 0.353 | her6 | -0.250 |
| kcnj1a.2 | 0.351 | calm1b | -0.250 |
| prlra | 0.350 | ankrd22 | -0.245 |