DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous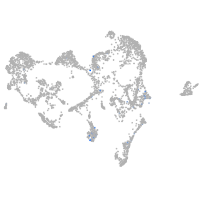 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dnd1 | 0.777 | ptmaa | -0.675 |
| nanos3 | 0.759 | h3f3a | -0.624 |
| nasp | 0.755 | ppiab | -0.597 |
| gra | 0.731 | si:ch211-191j22.3 | -0.578 |
| tdrd7a | 0.713 | wu:fb55g09 | -0.557 |
| stmn1a | 0.713 | calm1a | -0.555 |
| seta | 0.667 | pvalb1 | -0.547 |
| ca15b | 0.643 | anxa5b | -0.546 |
| ppig | 0.643 | scgn | -0.545 |
| snu13b | 0.637 | icn | -0.544 |
| surf2 | 0.632 | ucp2 | -0.542 |
| zgc:77118 | 0.629 | rpl37 | -0.541 |
| rpl7l1 | 0.615 | pvalb2 | -0.535 |
| thoc7 | 0.615 | faua | -0.533 |
| h1m | 0.613 | atp5f1b | -0.527 |
| pttg1 | 0.612 | cox8a | -0.523 |
| syncrip | 0.611 | ubc | -0.522 |
| rsl1d1 | 0.610 | h3f3c | -0.522 |
| bms1 | 0.593 | ccni | -0.520 |
| ddx18 | 0.581 | cyt1 | -0.519 |
| hnrnpa0b | 0.580 | syt1a | -0.515 |
| nop56 | 0.579 | ldhba | -0.514 |
| NC-002333.4 | 0.577 | rnasekb | -0.509 |
| ebna1bp2 | 0.575 | ier2b | -0.506 |
| kri1 | 0.572 | insm1b | -0.505 |
| dkc1 | 0.570 | rps17 | -0.502 |
| hnrnpabb | 0.567 | apoa2 | -0.502 |
| hnrnpub | 0.567 | mylpfa | -0.502 |
| si:dkey-67c22.2 | 0.566 | dap1b | -0.500 |
| gnl3 | 0.565 | CR383676.1 | -0.490 |
| slbp | 0.562 | atp6v0cb | -0.490 |
| akap12b | 0.558 | zgc:158463 | -0.488 |
| c1d | 0.558 | dpm2 | -0.488 |
| marcksl1b | 0.552 | b2ml | -0.486 |
| matr3l1.1 | 0.552 | CABZ01095001.1 | -0.486 |




