DEAD (Asp-Glu-Ala-Asp) box polypeptide 4
ZFIN 
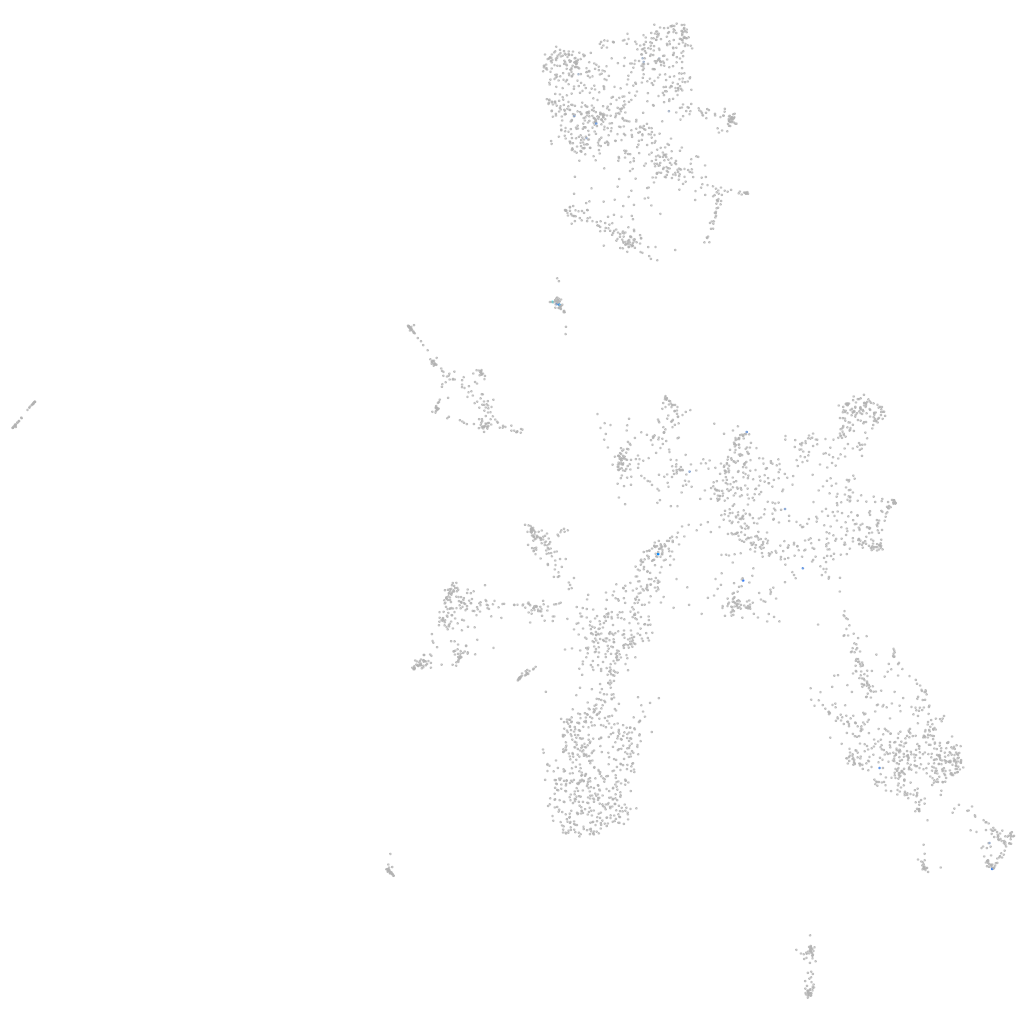
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm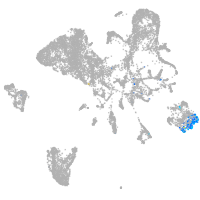 |
axial mesoderm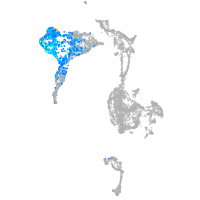 |
muscle 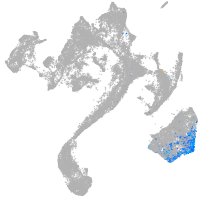 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 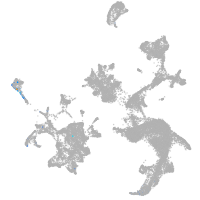 |
pronephros  |
neural |
spinal cord / glia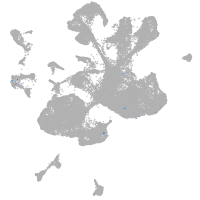 |
eye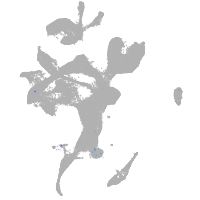 |
taste / olfactory |
otic / lateral line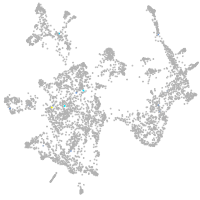 |
epidermis |
periderm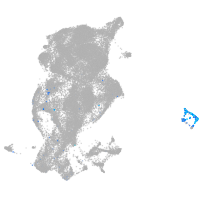 |
fin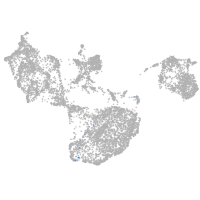 |
ionocytes / mucous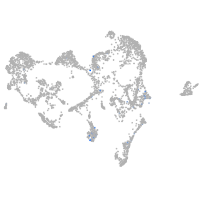 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| chia.3 | 0.629 | hmgn7 | -0.048 |
| ptk2ba | 0.364 | gapdhs | -0.045 |
| XLOC-028531 | 0.356 | atp1b1a | -0.043 |
| XLOC-001482 | 0.306 | tegt | -0.042 |
| nobox | 0.277 | calm2b | -0.042 |
| mslnb | 0.270 | tmsb4x | -0.041 |
| neto2a | 0.246 | oaz1b | -0.041 |
| reps2 | 0.245 | atp2b2 | -0.041 |
| LOC110437954 | 0.231 | atp6v0cb | -0.041 |
| cacna1sb | 0.211 | tspan2a | -0.040 |
| si:dkey-110g7.8 | 0.200 | uqcrh | -0.038 |
| si:dkey-98j1.5 | 0.197 | tuba1c | -0.038 |
| nod1 | 0.184 | sncb | -0.038 |
| tmc5 | 0.174 | zgc:65894 | -0.037 |
| rhoj | 0.173 | gnb1a | -0.037 |
| si:dkey-35i13.1 | 0.169 | gng3 | -0.037 |
| CT990625.1 | 0.169 | arpc1a | -0.036 |
| ficd | 0.163 | clstn1 | -0.036 |
| nansb | 0.160 | adcyap1b | -0.036 |
| stxbp6l | 0.156 | tmem258 | -0.036 |
| gla | 0.155 | ap2m1a | -0.036 |
| fam219b | 0.154 | serinc1 | -0.035 |
| GARNL3 | 0.151 | chga | -0.035 |
| tmtops2b | 0.150 | rtn1a | -0.035 |
| LO017850.1 | 0.149 | mt-nd2 | -0.035 |
| LOC103910668 | 0.147 | pvalb5 | -0.035 |
| slc22a23 | 0.147 | elavl3 | -0.035 |
| hivep3b | 0.145 | mycbp | -0.035 |
| ret | 0.145 | rnf10 | -0.035 |
| mkrn2os.1 | 0.143 | dap1b | -0.034 |
| tmprss4b | 0.139 | pim3 | -0.034 |
| npy2r | 0.137 | atf5a | -0.034 |
| rce1a | 0.136 | dynll1 | -0.034 |
| znf1007 | 0.135 | hmgb1b | -0.034 |
| nedd4a | 0.134 | ssr4 | -0.034 |