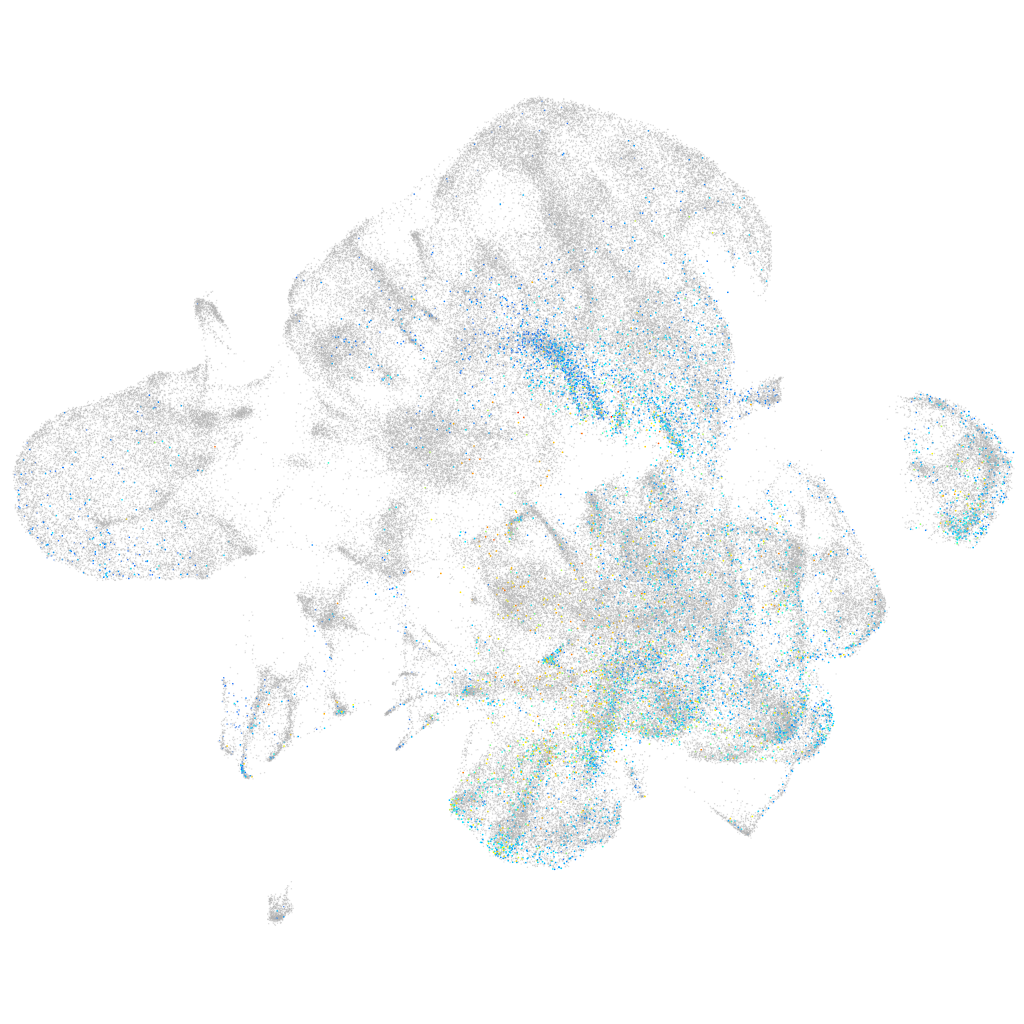
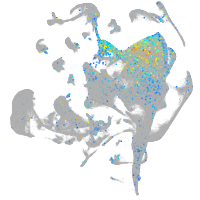
CUE domain containing 1a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rtn1a | 0.121 | id1 | -0.098 |
| rnasekb | 0.119 | nop58 | -0.095 |
| gapdhs | 0.118 | npm1a | -0.094 |
| ckbb | 0.117 | pcna | -0.088 |
| vamp2 | 0.116 | snu13b | -0.086 |
| stmn1b | 0.115 | nop56 | -0.085 |
| atp6v1g1 | 0.114 | dkc1 | -0.085 |
| atp6v0cb | 0.114 | hmgb2a | -0.084 |
| atp6v1e1b | 0.114 | fbl | -0.083 |
| tpi1b | 0.113 | si:dkey-151g10.6 | -0.082 |
| ywhag2 | 0.109 | rsl1d1 | -0.080 |
| atpv0e2 | 0.108 | nop10 | -0.078 |
| necap1 | 0.108 | mcm6 | -0.077 |
| gpm6aa | 0.107 | sdc4 | -0.077 |
| tuba1c | 0.106 | ranbp1 | -0.077 |
| si:dkeyp-75h12.5 | 0.105 | selenoh | -0.077 |
| sncb | 0.103 | rplp1 | -0.076 |
| eno2 | 0.103 | nhp2 | -0.076 |
| gng3 | 0.102 | chaf1a | -0.075 |
| snap25a | 0.102 | hmga1a | -0.074 |
| sh3gl2a | 0.101 | lig1 | -0.074 |
| zgc:65894 | 0.101 | nop2 | -0.074 |
| elavl3 | 0.101 | cdca7b | -0.073 |
| calm1a | 0.101 | mcm2 | -0.073 |
| sypb | 0.101 | anp32b | -0.072 |
| sncgb | 0.101 | paics | -0.072 |
| ndufa4 | 0.099 | si:ch211-217k17.7 | -0.072 |
| eno1a | 0.099 | si:dkey-102m7.3 | -0.071 |
| stx1b | 0.099 | rps28 | -0.071 |
| calm1b | 0.099 | akap12b | -0.071 |
| stxbp1a | 0.098 | nasp | -0.071 |
| si:ch73-119p20.1 | 0.098 | rps12 | -0.070 |
| gpm6ab | 0.097 | mcm3 | -0.070 |
| snap25b | 0.097 | mcm5 | -0.070 |
| fez1 | 0.097 | rplp2l | -0.070 |