cryptochrome circadian regulator 1aa [Source:ZFIN;Acc:ZDB-GENE-010426-2]
ZFIN 
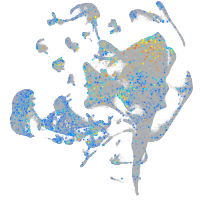
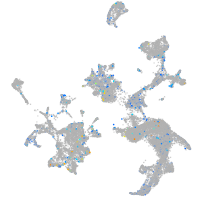
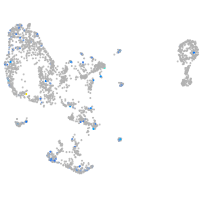
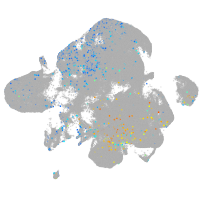
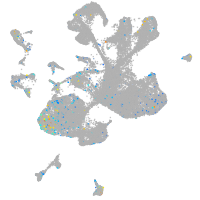
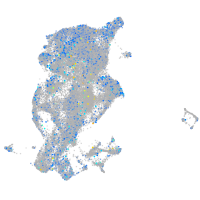
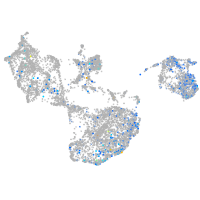
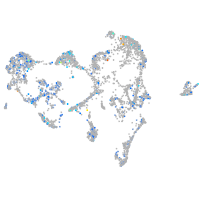
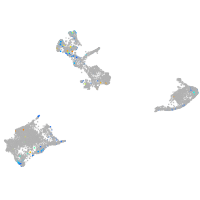
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC108190423 | 0.071 | cirbpb | -0.064 |
| oaz2b | 0.056 | si:ch211-222l21.1 | -0.062 |
| aldocb | 0.054 | ptmab | -0.058 |
| egr4 | 0.053 | hnrnpaba | -0.054 |
| fosab | 0.051 | khdrbs1a | -0.053 |
| sncgb | 0.051 | si:dkey-56m19.5 | -0.051 |
| prss1 | 0.050 | ybx1 | -0.051 |
| bhlhe40 | 0.050 | tubb2b | -0.050 |
| bhlhe41 | 0.050 | hmgn6 | -0.049 |
| ier2b | 0.048 | chd4a | -0.048 |
| eno1a | 0.047 | tubb5 | -0.047 |
| CU062647.1 | 0.047 | hnrnpa0b | -0.046 |
| si:ch211-107e6.6 | 0.046 | sox11b | -0.045 |
| apoa1b | 0.045 | rps12 | -0.045 |
| prss59.2 | 0.045 | hnrnpa0a | -0.045 |
| pkma | 0.045 | tuba1a | -0.044 |
| slc6a17 | 0.045 | hmga1a | -0.044 |
| npas4a | 0.044 | eef1a1l1 | -0.044 |
| gapdhs | 0.044 | hnrnpabb | -0.043 |
| atp1b1b | 0.043 | si:ch211-288g17.3 | -0.043 |
| mir132-2 | 0.043 | tpt1 | -0.043 |
| vsnl1b | 0.043 | hsp90ab1 | -0.042 |
| calm1b | 0.042 | rpsa | -0.042 |
| stmn4 | 0.042 | rplp2l | -0.040 |
| eno2 | 0.042 | hdac1 | -0.040 |
| tob1a | 0.041 | tmeff1b | -0.040 |
| glula | 0.041 | tmsb | -0.039 |
| nupr1a | 0.041 | hmgn2 | -0.038 |
| atp2b2 | 0.041 | rpl7a | -0.038 |
| tefb | 0.040 | rps20 | -0.037 |
| gchfr | 0.040 | rps16 | -0.037 |
| apoa2 | 0.040 | nono | -0.036 |
| snap25a | 0.039 | rps8a | -0.036 |
| sod2 | 0.039 | marcksb | -0.036 |
| camk2n1a | 0.039 | actb1 | -0.036 |