"complexin 2, like"
ZFIN 
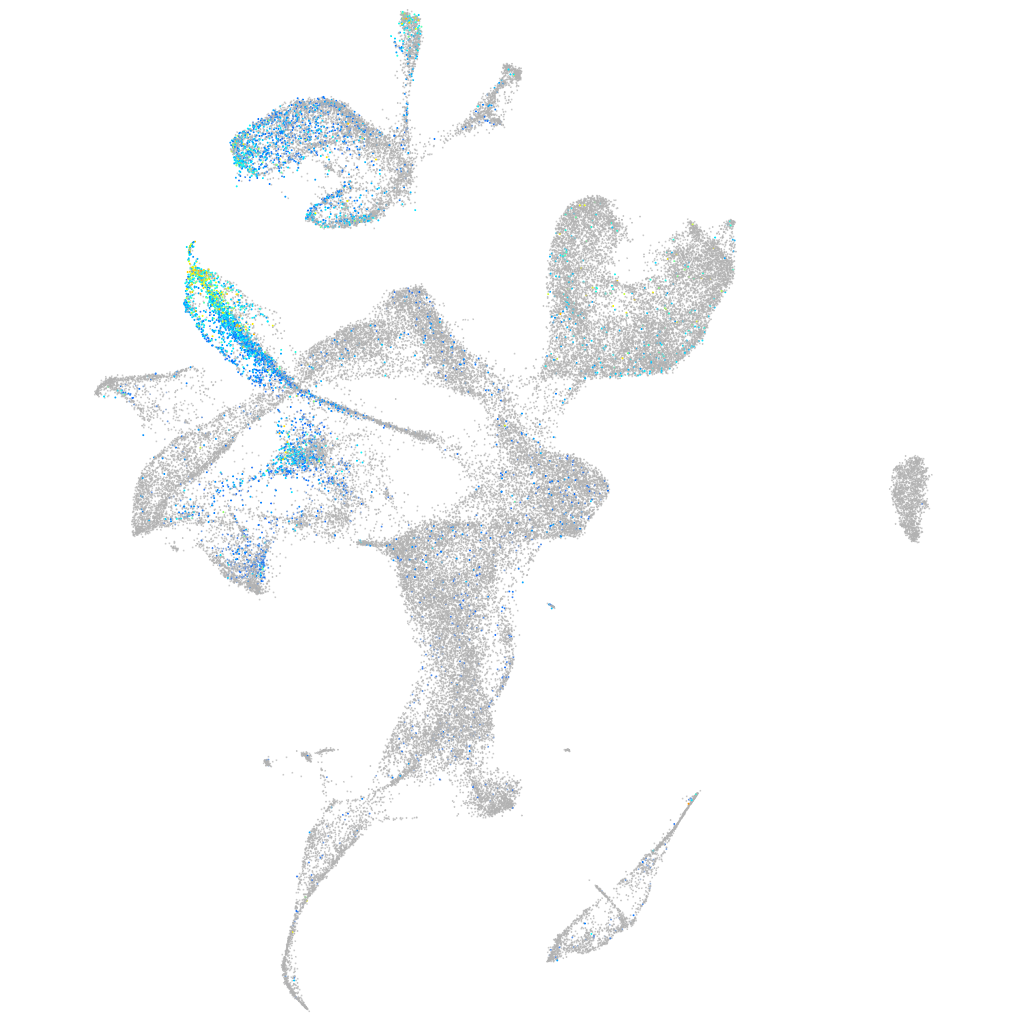
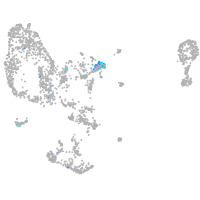
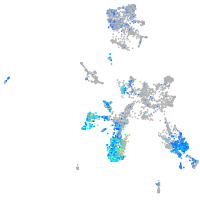
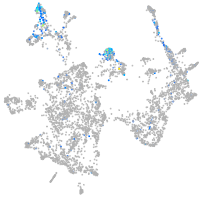
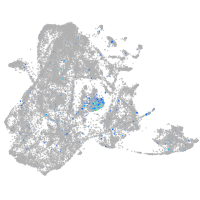
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rbpms2a | 0.471 | hmgb2a | -0.262 |
| eno2 | 0.440 | hmga1a | -0.144 |
| syt2a | 0.421 | mdka | -0.141 |
| stmn2a | 0.413 | pcna | -0.140 |
| isl2b | 0.407 | stmn1a | -0.139 |
| inab | 0.407 | fabp7a | -0.135 |
| rtn1b | 0.404 | msi1 | -0.135 |
| rbpms2b | 0.400 | ahcy | -0.135 |
| pou4f1 | 0.389 | nutf2l | -0.130 |
| snap25a | 0.385 | rrm1 | -0.129 |
| zgc:65894 | 0.382 | ccnd1 | -0.128 |
| ywhag2 | 0.380 | dut | -0.127 |
| islr2 | 0.377 | msna | -0.126 |
| oaz2b | 0.372 | mki67 | -0.126 |
| gng3 | 0.364 | mcm7 | -0.124 |
| stxbp1a | 0.360 | chaf1a | -0.124 |
| elavl3 | 0.351 | lbr | -0.122 |
| calb2a | 0.351 | selenoh | -0.120 |
| rbfox1 | 0.349 | tuba8l | -0.118 |
| BX247868.1 | 0.348 | eef1da | -0.118 |
| zgc:153426 | 0.348 | COX7A2 (1 of many) | -0.117 |
| zgc:158291 | 0.347 | her15.1 | -0.116 |
| grin1a | 0.346 | ccna2 | -0.116 |
| vsnl1b | 0.343 | rpa3 | -0.114 |
| id4 | 0.343 | dek | -0.113 |
| stmn4l | 0.342 | lig1 | -0.113 |
| sprn | 0.340 | banf1 | -0.112 |
| atp2b3b | 0.338 | id1 | -0.110 |
| stx1b | 0.335 | hmgb2b | -0.109 |
| nrn1a | 0.333 | rps20 | -0.108 |
| pou4f2 | 0.332 | cks1b | -0.108 |
| stmn4 | 0.332 | cx43.4 | -0.108 |
| pou4f3 | 0.332 | mcm6 | -0.107 |
| mllt11 | 0.331 | rrm2 | -0.106 |
| cplx2 | 0.325 | fen1 | -0.106 |