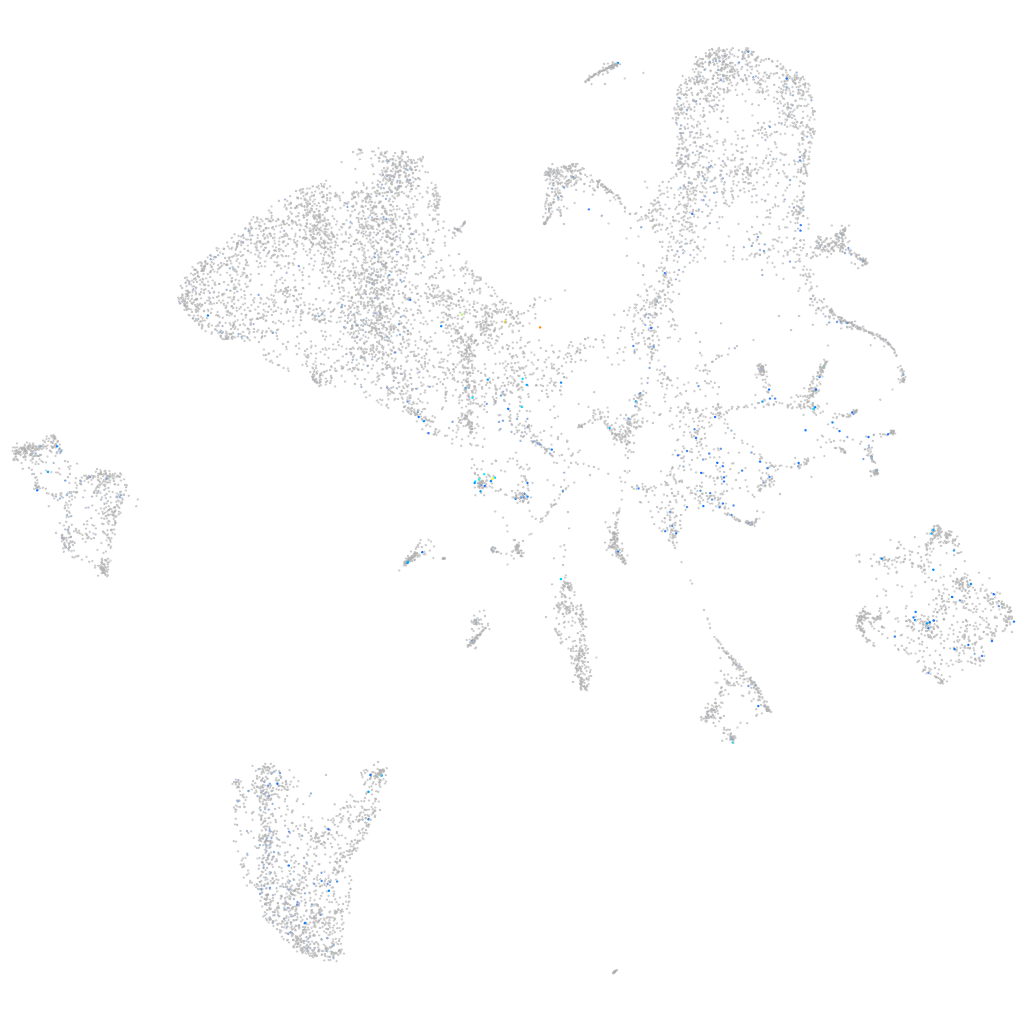
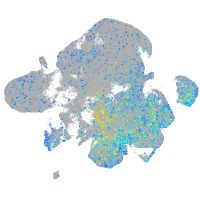
claudin domain containing 1a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101884310 | 0.172 | aldob | -0.067 |
| si:ch211-239f4.1 | 0.152 | gapdh | -0.064 |
| CABZ01118575.1 | 0.136 | eno3 | -0.064 |
| CU929294.1 | 0.133 | apoa4b.1 | -0.062 |
| CU896691.1 | 0.130 | gpx4a | -0.061 |
| rx2 | 0.129 | gamt | -0.060 |
| ankrd6a | 0.128 | afp4 | -0.060 |
| lyve1a | 0.127 | scp2a | -0.059 |
| rorb | 0.125 | apoc2 | -0.058 |
| si:ch211-57b15.2 | 0.125 | pklr | -0.057 |
| zgc:91999 | 0.119 | fbp1b | -0.057 |
| pimr197 | 0.119 | ces2 | -0.057 |
| CR407590.2 | 0.118 | rbp4 | -0.057 |
| XLOC-019975 | 0.117 | apom | -0.056 |
| LOC101883911 | 0.115 | sult2st2 | -0.055 |
| LOC100332354 | 0.114 | apoa1b | -0.055 |
| si:ch211-266k22.6 | 0.112 | hao1 | -0.055 |
| clstn3 | 0.112 | ckba | -0.055 |
| bcan | 0.112 | gc | -0.055 |
| rrh | 0.111 | mgst1.2 | -0.054 |
| LOC103908684 | 0.110 | suclg1 | -0.054 |
| si:ch211-286b4.4 | 0.110 | pgk1 | -0.053 |
| kcnh2a | 0.110 | ttr | -0.053 |
| cnrip1b | 0.110 | pgm1 | -0.053 |
| cadm3 | 0.109 | rgrb | -0.053 |
| LOC108192008 | 0.106 | itih2 | -0.052 |
| neurod4 | 0.105 | gstt1a | -0.052 |
| LOC103911092 | 0.105 | tfa | -0.052 |
| si:ch211-284e13.6 | 0.103 | apobb.1 | -0.052 |
| XLOC-044186 | 0.103 | rdh1 | -0.052 |
| mthfd2l | 0.102 | c9 | -0.052 |
| prickle2a | 0.102 | ambp | -0.052 |
| ldb2b | 0.101 | grhprb | -0.052 |
| AL954831.1 | 0.101 | etnppl | -0.051 |
| LOC100329307 | 0.101 | comtd1 | -0.051 |