chondroitin sulfate synthase 1
ZFIN 
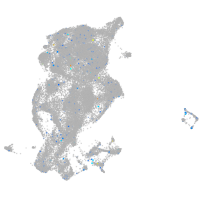
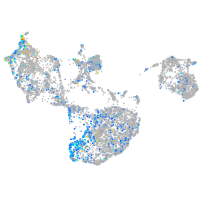
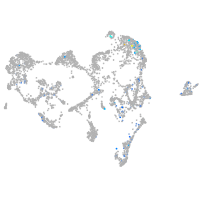
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| col8a1a | 0.155 | mdkb | -0.056 |
| aplnr2 | 0.143 | ckbb | -0.048 |
| sulf1 | 0.134 | slc1a2b | -0.048 |
| col11a1a | 0.133 | CR848047.1 | -0.041 |
| plod2 | 0.127 | zgc:165461 | -0.041 |
| optc | 0.124 | CU467822.1 | -0.040 |
| col9a3 | 0.123 | fabp7a | -0.040 |
| col9a2 | 0.118 | efhd1 | -0.040 |
| spon2a | 0.117 | mdka | -0.039 |
| cyr61l2 | 0.116 | crabp1a | -0.037 |
| ctgfa | 0.116 | glula | -0.036 |
| oc90 | 0.115 | acbd7 | -0.035 |
| foxa | 0.113 | ppap2d | -0.035 |
| slit1b | 0.108 | tmsb4x | -0.033 |
| col2a1a | 0.108 | pvalb1 | -0.031 |
| col5a1 | 0.104 | slc4a4a | -0.031 |
| BX908782.3 | 0.103 | hbbe1.3 | -0.031 |
| spon1a | 0.102 | nfia | -0.031 |
| col9a1b | 0.102 | atp1a1b | -0.030 |
| spon1b | 0.101 | hbae3 | -0.030 |
| ntn1b | 0.098 | mt2 | -0.030 |
| shhb | 0.096 | pvalb2 | -0.030 |
| myl9a | 0.095 | syngr2b | -0.030 |
| npr3 | 0.094 | gas1b | -0.029 |
| hapln1a | 0.093 | ca4a | -0.028 |
| cers3b | 0.093 | gpm6ab | -0.028 |
| col2a1b | 0.092 | cahz | -0.028 |
| tbata | 0.091 | nrgna | -0.027 |
| shha | 0.091 | ndrg3a | -0.027 |
| foxa2 | 0.091 | cx43 | -0.027 |
| clu | 0.091 | slc1a3b | -0.027 |
| loxl5b | 0.090 | hbbe1.2 | -0.026 |
| scospondin | 0.089 | stmn1b | -0.026 |
| FP102018.1 | 0.089 | cspg5a | -0.026 |
| chl1a | 0.089 | apoa1b | -0.026 |