charged multivesicular body protein 5a
ZFIN 
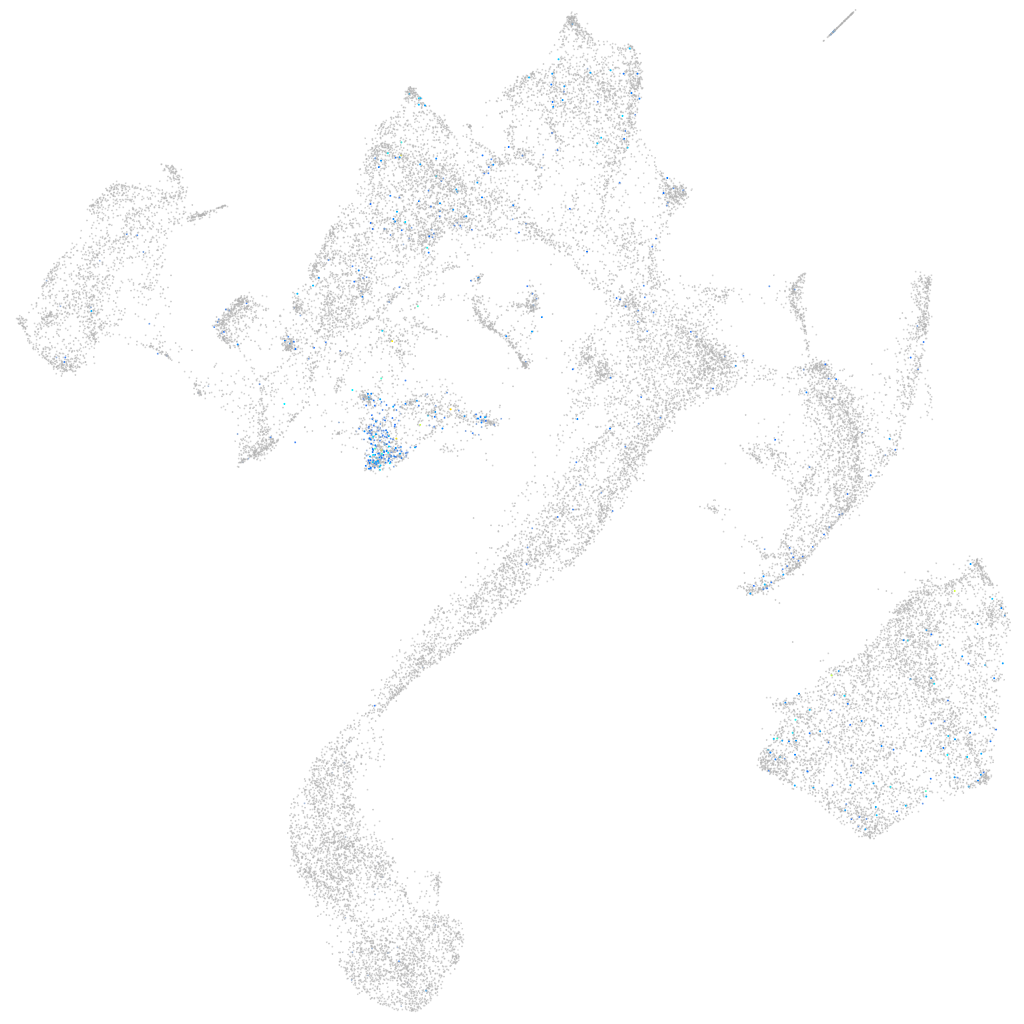
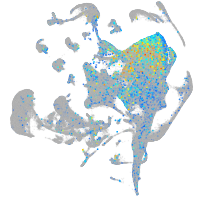
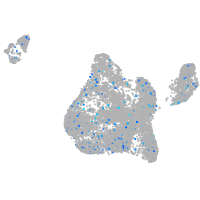
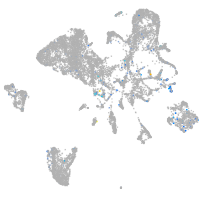
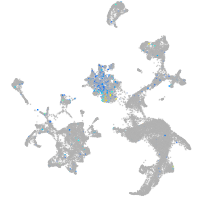
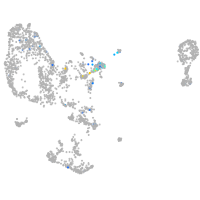
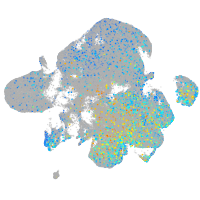
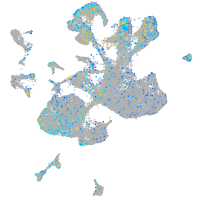
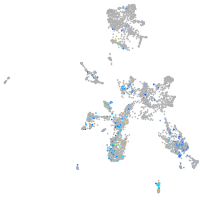
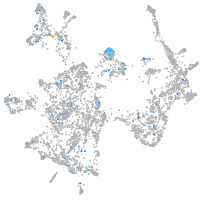
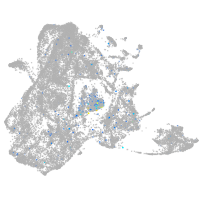
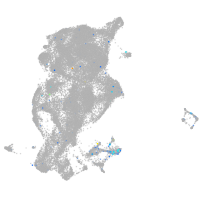
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpm6aa | 0.198 | tuba8l2 | -0.061 |
| stmn1b | 0.184 | ttn.1 | -0.060 |
| gng3 | 0.183 | srl | -0.054 |
| sncb | 0.176 | aldoab | -0.054 |
| rtn1b | 0.173 | ttn.2 | -0.054 |
| tuba1c | 0.169 | tmem38a | -0.054 |
| stxbp1a | 0.168 | mybphb | -0.053 |
| atp1a3a | 0.168 | ldb3a | -0.053 |
| stx1b | 0.167 | zgc:92429 | -0.053 |
| ppp1r14ba | 0.166 | zgc:101853 | -0.052 |
| stmn2a | 0.165 | tpma | -0.052 |
| elavl4 | 0.164 | CABZ01078594.1 | -0.052 |
| vamp2 | 0.162 | atp2a1 | -0.051 |
| syt1a | 0.161 | CABZ01072309.1 | -0.051 |
| olfm1b | 0.160 | hsp90aa1.1 | -0.050 |
| elavl3 | 0.160 | tnnc2 | -0.050 |
| zgc:153426 | 0.160 | unc45b | -0.050 |
| fez1 | 0.155 | actc1a | -0.050 |
| ckbb | 0.153 | acta1b | -0.050 |
| cspg5a | 0.152 | neb | -0.049 |
| zgc:65894 | 0.151 | txlnbb | -0.049 |
| eno2 | 0.151 | desma | -0.049 |
| cnrip1a | 0.149 | gamt | -0.049 |
| rnasekb | 0.148 | actc1b | -0.049 |
| snap25a | 0.148 | ckma | -0.049 |
| gabrb4 | 0.148 | klhl31 | -0.049 |
| nsfa | 0.147 | acta1a | -0.048 |
| snap25b | 0.147 | actn3b | -0.048 |
| slitrk1 | 0.147 | gapdh | -0.048 |
| fam117ba | 0.143 | tnni2b.1 | -0.048 |
| gap43 | 0.143 | atp1a2a | -0.047 |
| gnao1a | 0.143 | ak1 | -0.047 |
| sv2a | 0.142 | BX276101.1 | -0.047 |
| sypa | 0.142 | ryr1b | -0.047 |
| myt1la | 0.142 | cav3 | -0.047 |