cilia and flagella associated protein 300
ZFIN 
Other cell groups


all cells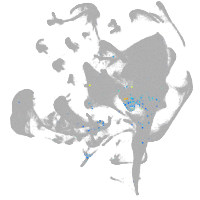 |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia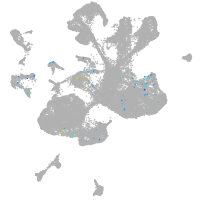 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110438335 | 0.113 | elavl3 | -0.036 |
| daw1 | 0.112 | stmn1b | -0.033 |
| tbata | 0.107 | myt1b | -0.031 |
| si:dkey-148f10.4 | 0.103 | gng3 | -0.031 |
| efcab1 | 0.101 | rtn1a | -0.030 |
| si:ch211-155m12.5 | 0.100 | si:dkey-276j7.1 | -0.029 |
| enkur | 0.098 | epb41a | -0.028 |
| ccdc114 | 0.097 | stxbp1a | -0.028 |
| si:ch211-248e11.2 | 0.096 | elavl4 | -0.027 |
| foxj1a | 0.095 | stx1b | -0.027 |
| ttc25 | 0.093 | atp6v0cb | -0.026 |
| LOC103910155 | 0.093 | ywhah | -0.026 |
| ribc2 | 0.092 | golga7ba | -0.026 |
| smkr1 | 0.090 | atp1b2a | -0.026 |
| LOC103910742 | 0.090 | scrt2 | -0.025 |
| ccdc173 | 0.089 | elovl4a | -0.025 |
| lrrc6 | 0.088 | aplp1 | -0.025 |
| cfap57 | 0.087 | nsg2 | -0.025 |
| rab36 | 0.086 | vamp2 | -0.024 |
| pacrg | 0.085 | onecut1 | -0.024 |
| zgc:55461 | 0.085 | cplx2 | -0.024 |
| cfap126 | 0.084 | rtn1b | -0.024 |
| drc1 | 0.084 | ptmaa | -0.024 |
| si:ch211-226m7.4 | 0.084 | nhlh2 | -0.024 |
| catip | 0.083 | cdk5r1b | -0.024 |
| tppp3 | 0.083 | tmem59l | -0.024 |
| ak7a | 0.082 | tmsb | -0.023 |
| ccdc169 | 0.082 | myt1a | -0.023 |
| efhc1 | 0.082 | si:ch73-386h18.1 | -0.023 |
| FP016142.4 | 0.081 | ndrg4 | -0.023 |
| meig1 | 0.081 | stmn2a | -0.023 |
| dydc2 | 0.080 | srrm4 | -0.023 |
| krt18a.1 | 0.079 | snap25a | -0.023 |
| LOC103911747 | 0.079 | necap1 | -0.023 |
| cfap157 | 0.079 | ywhag2 | -0.022 |




