centromere protein S
ZFIN 
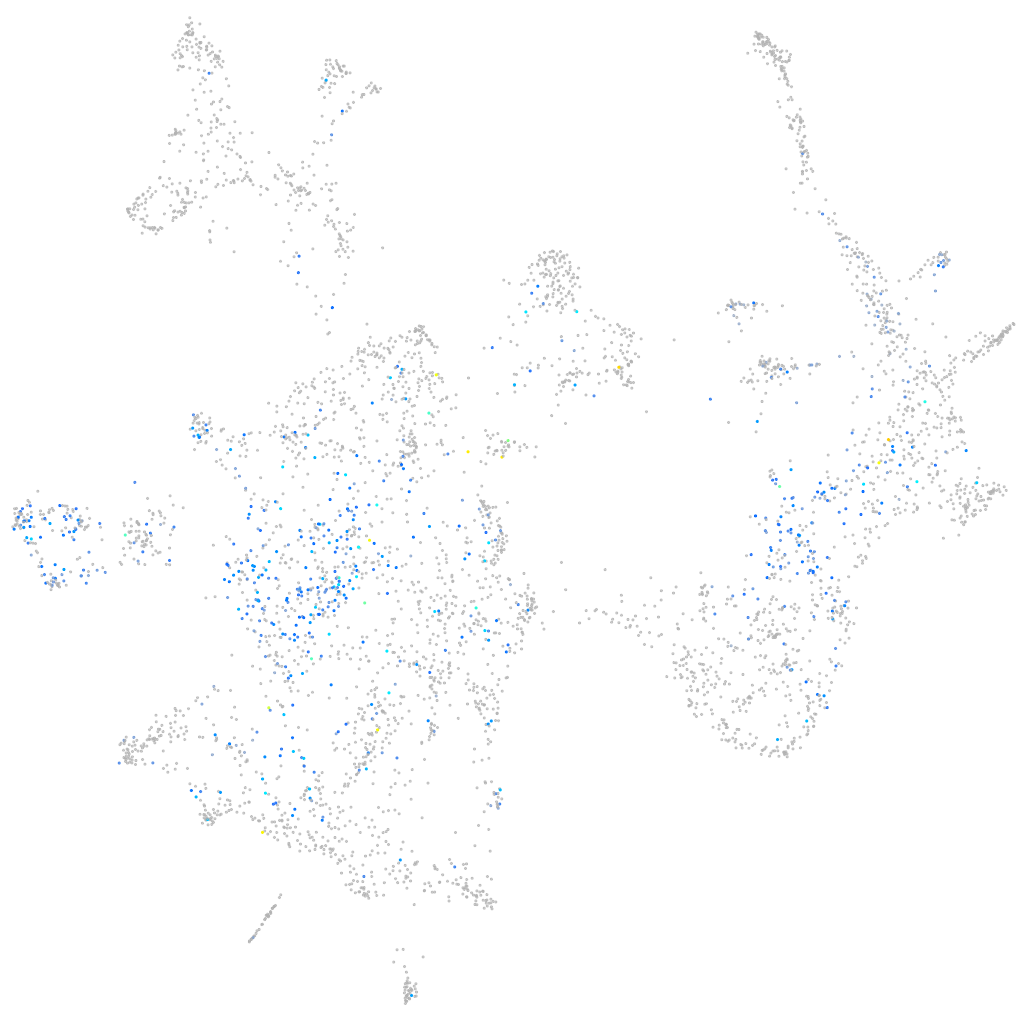
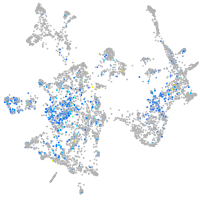
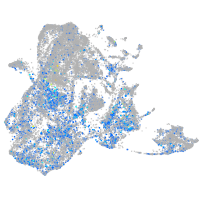
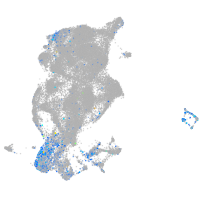
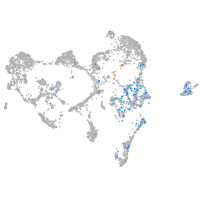
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dut | 0.405 | tob1b | -0.145 |
| stmn1a | 0.392 | nupr1a | -0.129 |
| pcna | 0.377 | h1f0 | -0.129 |
| rrm1 | 0.365 | rnasekb | -0.128 |
| cks1b | 0.356 | CR383676.1 | -0.125 |
| rpa3 | 0.348 | ccni | -0.125 |
| chaf1a | 0.348 | calm1b | -0.122 |
| si:dkey-6i22.5 | 0.347 | tmem59 | -0.119 |
| dhfr | 0.340 | atp6v1e1b | -0.119 |
| aurkb | 0.334 | gabarapl2 | -0.118 |
| tubb2b | 0.334 | nptna | -0.117 |
| hmgb2a | 0.331 | kif1aa | -0.117 |
| mki67 | 0.331 | cd164l2 | -0.116 |
| anp32b | 0.331 | otofb | -0.116 |
| nasp | 0.329 | gpx2 | -0.115 |
| banf1 | 0.328 | osbpl1a | -0.115 |
| cdk1 | 0.327 | dnajc5b | -0.114 |
| cdca5 | 0.324 | calm1a | -0.114 |
| mad2l1 | 0.324 | pvalb8 | -0.113 |
| rpa2 | 0.322 | pkig | -0.113 |
| zgc:110540 | 0.322 | CR925719.1 | -0.113 |
| ccna2 | 0.317 | aldocb | -0.112 |
| rrm2 | 0.316 | gapdhs | -0.112 |
| si:ch211-156b7.4 | 0.310 | eno1a | -0.112 |
| spc25 | 0.310 | mcl1a | -0.112 |
| tyms | 0.310 | ckbb | -0.112 |
| hmgb2b | 0.305 | atp2b1a | -0.111 |
| mibp | 0.303 | emb | -0.109 |
| smc2 | 0.303 | atp1a3b | -0.109 |
| LOC100330864 | 0.302 | calml4a | -0.109 |
| dek | 0.302 | prr15la | -0.108 |
| top2a | 0.302 | gabarapa | -0.108 |
| fen1 | 0.301 | ap3s1.1 | -0.107 |
| dlgap5 | 0.300 | si:ch73-15n15.3 | -0.107 |
| lig1 | 0.300 | atp5if1b | -0.106 |