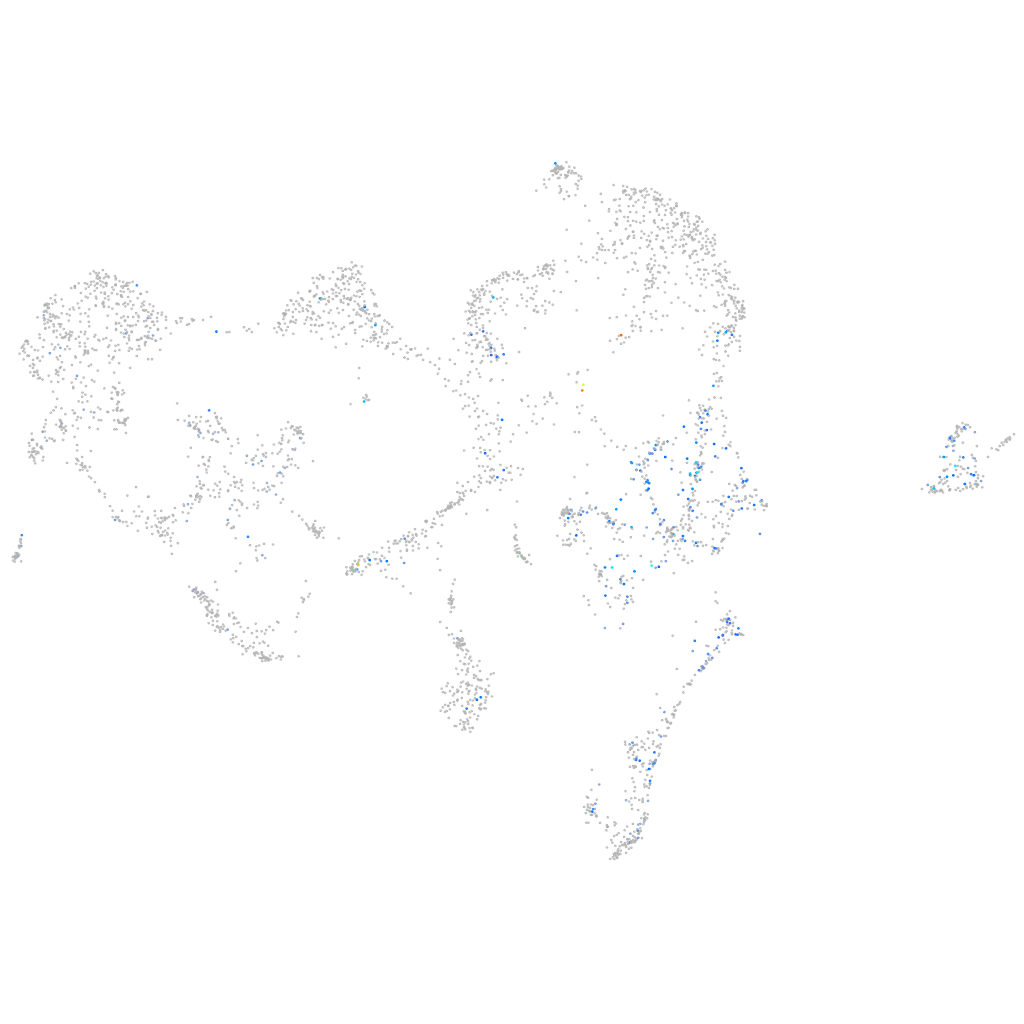
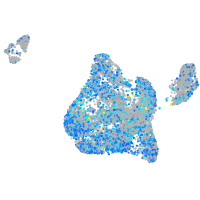
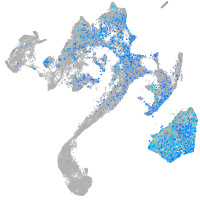
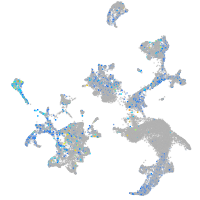
centromere protein S
ZFIN 
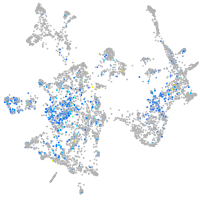
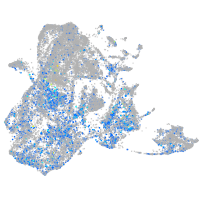
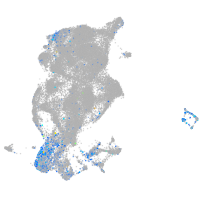
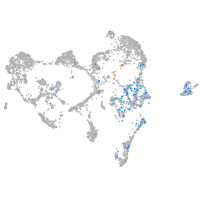
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dut | 0.357 | selenow1 | -0.162 |
| si:dkey-261m9.17 | 0.316 | si:ch211-195b11.3 | -0.144 |
| zgc:110540 | 0.292 | aldob | -0.132 |
| stmn1a | 0.290 | BX000438.2 | -0.130 |
| fbxo5 | 0.285 | si:dkey-193p11.2 | -0.127 |
| rrm1 | 0.278 | tob1b | -0.122 |
| pcna | 0.270 | zgc:193505 | -0.122 |
| cks1b | 0.268 | abcb5 | -0.121 |
| hmgb2b | 0.267 | icn2 | -0.119 |
| tyms | 0.267 | selenow2b | -0.115 |
| smc2 | 0.265 | si:dkey-248g15.3 | -0.114 |
| si:ch211-288g17.3 | 0.261 | epdl1 | -0.113 |
| rpa2 | 0.258 | si:dkey-247k7.2 | -0.111 |
| dek | 0.255 | cst14b.1 | -0.110 |
| tubb2b | 0.255 | zgc:153284 | -0.110 |
| ncapd2 | 0.254 | krt4 | -0.110 |
| FO744840.1 | 0.253 | mgst1.2 | -0.109 |
| mki67 | 0.252 | nupr1a | -0.109 |
| smc4 | 0.252 | wu:fb18f06 | -0.109 |
| rpa3 | 0.248 | fbp1a | -0.108 |
| chaf1a | 0.246 | sqstm1 | -0.108 |
| cdk1 | 0.246 | mt-co1 | -0.107 |
| dhfr | 0.244 | scel | -0.106 |
| cenpx | 0.244 | enosf1 | -0.106 |
| aurkb | 0.242 | mt-cyb | -0.105 |
| LOC108181115 | 0.242 | tdh | -0.104 |
| rbbp4 | 0.240 | soul2 | -0.103 |
| mis12 | 0.239 | nfe2l2a | -0.103 |
| ccna2 | 0.238 | mt-co2 | -0.103 |
| seta | 0.237 | ier2b | -0.103 |
| rrm2 | 0.237 | rplp2 | -0.103 |
| hmga1a | 0.233 | itm2bb | -0.102 |
| zgc:165555.3 | 0.232 | krt17 | -0.100 |
| cdca5 | 0.232 | CR383676.1 | -0.100 |
| anp32b | 0.232 | ppl | -0.099 |