"calmodulin 3b (phosphorylase kinase, delta)"
ZFIN 
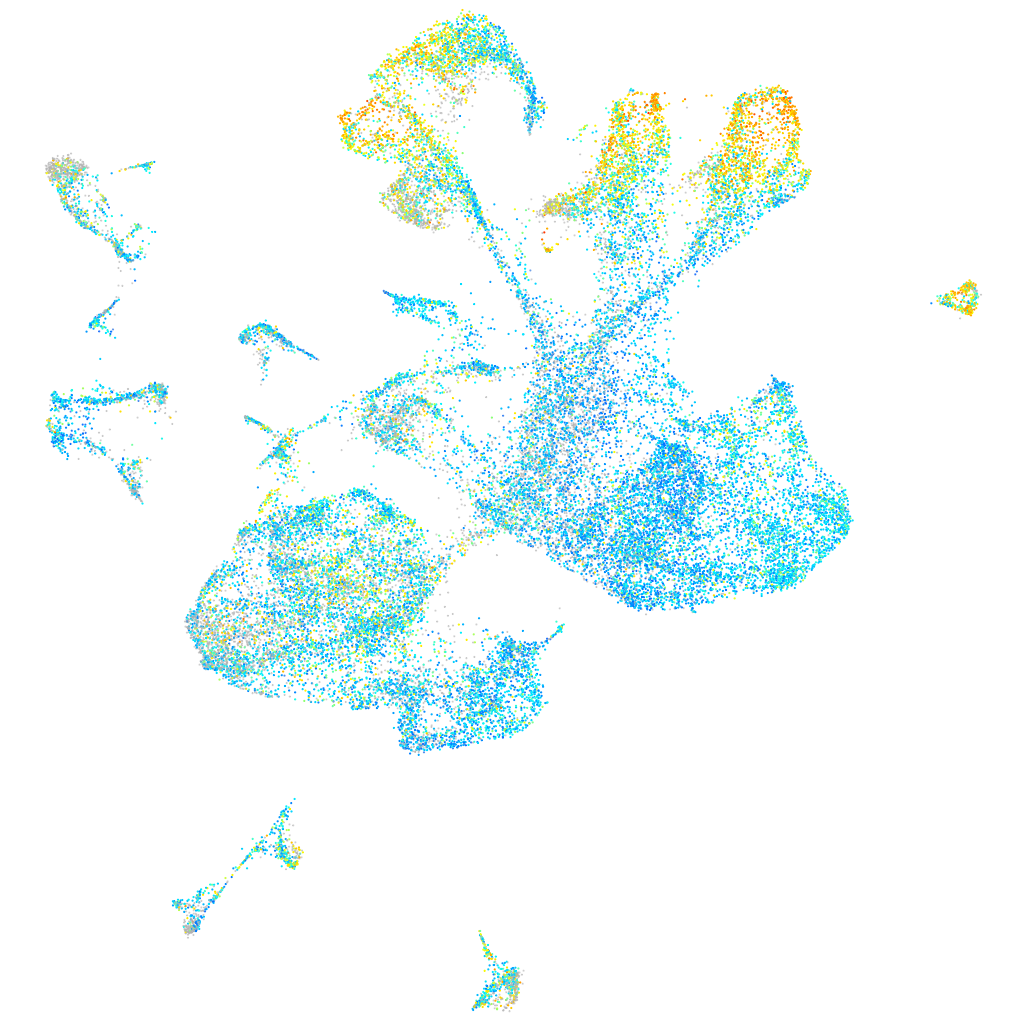
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ywhag2 | 0.362 | atp1a1b | -0.199 |
| gng3 | 0.347 | mdka | -0.191 |
| snap25a | 0.341 | sparc | -0.189 |
| sncb | 0.337 | gpm6bb | -0.170 |
| calm1b | 0.332 | notch3 | -0.169 |
| stmn2a | 0.330 | id1 | -0.153 |
| cplx2l | 0.329 | si:ch211-251b21.1 | -0.153 |
| stxbp1a | 0.327 | si:ch73-21g5.7 | -0.151 |
| zgc:65894 | 0.327 | gfap | -0.149 |
| calm2a | 0.324 | cldn19 | -0.147 |
| tubb2b | 0.324 | slc6a9 | -0.142 |
| rtn1b | 0.322 | cd63 | -0.142 |
| stx1b | 0.322 | si:ch211-57n23.4 | -0.141 |
| calm2b | 0.320 | qki2 | -0.137 |
| atp6v0cb | 0.318 | mir219-3 | -0.137 |
| tuba1c | 0.318 | zgc:165461 | -0.137 |
| sncgb | 0.318 | LOC798783 | -0.136 |
| sypb | 0.303 | slc1a3a | -0.136 |
| ndufa4 | 0.300 | gadd45gb.1 | -0.133 |
| atp6v1e1b | 0.300 | dla | -0.130 |
| syn2a | 0.296 | ptn | -0.130 |
| eno2 | 0.294 | fabp7a | -0.127 |
| atp5mc1 | 0.292 | sb:cb81 | -0.127 |
| vamp2 | 0.291 | XLOC-003690 | -0.124 |
| cnrip1a | 0.290 | cx43 | -0.122 |
| atpv0e2 | 0.290 | efcc1 | -0.120 |
| atp6v1g1 | 0.290 | btg2 | -0.120 |
| mllt11 | 0.286 | atp1b4 | -0.119 |
| ppiab | 0.284 | notch1a | -0.118 |
| zgc:153426 | 0.284 | lfng | -0.116 |
| nptna | 0.284 | nrarpb | -0.115 |
| sh3gl2a | 0.284 | fstl1a | -0.115 |
| id4 | 0.281 | slc4a4a | -0.115 |
| rbfox1 | 0.281 | si:dkey-42i9.4 | -0.114 |
| cplx2 | 0.280 | dld | -0.113 |