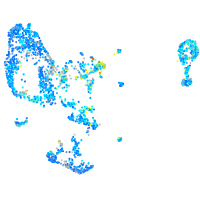"calmodulin 2b, (phosphorylase kinase, delta)"
ZFIN 
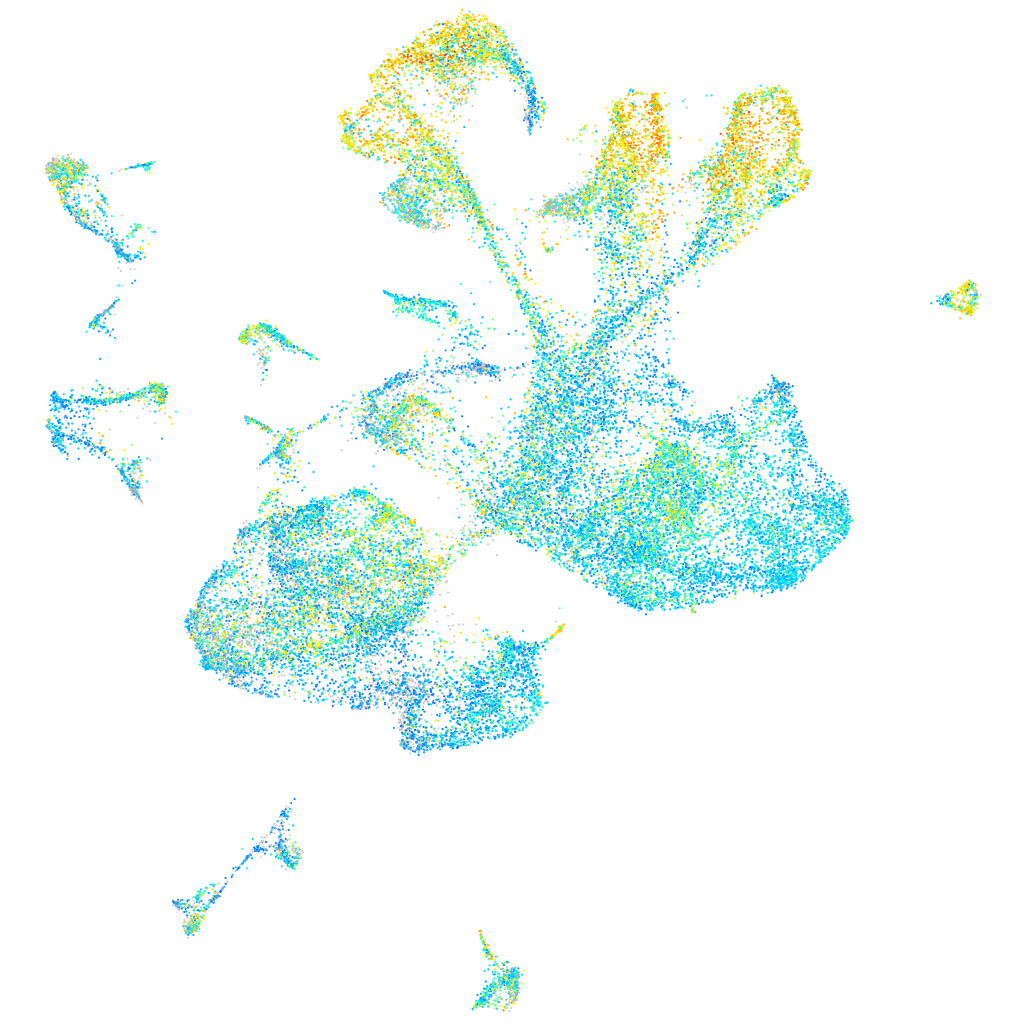
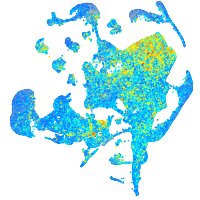
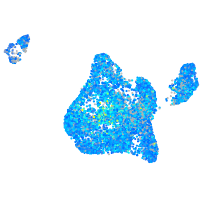
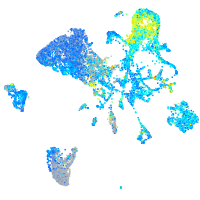
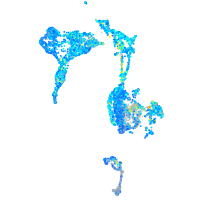
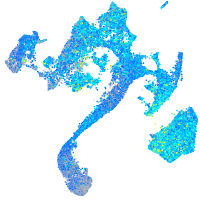
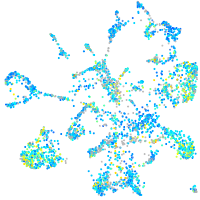
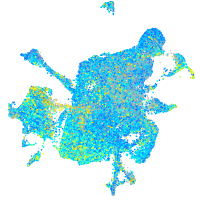
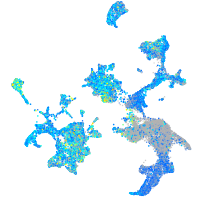
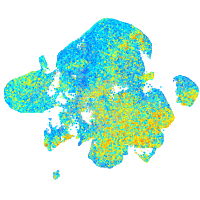
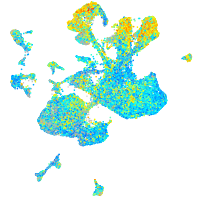
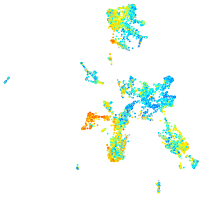
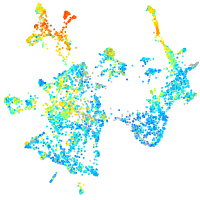
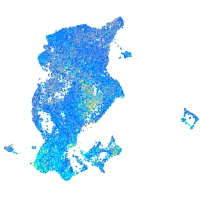
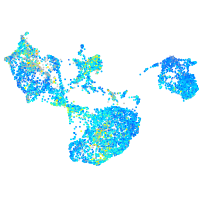
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sncb | 0.361 | atp1a1b | -0.218 |
| ywhag2 | 0.360 | gpm6bb | -0.199 |
| gng3 | 0.358 | fabp7a | -0.194 |
| snap25a | 0.344 | sparc | -0.193 |
| stxbp1a | 0.342 | ptn | -0.185 |
| stmn2a | 0.339 | cd63 | -0.176 |
| stx1b | 0.333 | XLOC-003690 | -0.175 |
| tuba1c | 0.333 | ntn1b | -0.166 |
| tubb2b | 0.333 | notch3 | -0.165 |
| cplx2l | 0.329 | nkx2.2a | -0.165 |
| cplx2 | 0.328 | fgfr2 | -0.162 |
| atp6v0cb | 0.327 | atp1b4 | -0.160 |
| calm3b | 0.320 | id1 | -0.159 |
| zgc:65894 | 0.316 | nkx2.2b | -0.157 |
| rtn1b | 0.313 | slc4a4a | -0.154 |
| calm2a | 0.312 | qki2 | -0.149 |
| vamp2 | 0.308 | slc6a9 | -0.148 |
| gap43 | 0.306 | cd82a | -0.146 |
| syt2a | 0.304 | ptgdsb.2 | -0.146 |
| sncgb | 0.303 | ptch2 | -0.146 |
| calm1b | 0.300 | CU467822.1 | -0.145 |
| elavl4 | 0.299 | sdc4 | -0.143 |
| cnrip1a | 0.295 | btg2 | -0.143 |
| atp6v1e1b | 0.292 | cx43 | -0.142 |
| eno2 | 0.288 | zgc:165461 | -0.141 |
| gng2 | 0.287 | si:ch211-66e2.5 | -0.138 |
| mllt11 | 0.285 | glula | -0.136 |
| atp1a3a | 0.281 | ppap2d | -0.135 |
| rbfox1 | 0.281 | slc6a11b | -0.133 |
| tmem59l | 0.280 | mfge8a | -0.132 |
| si:dkey-276j7.1 | 0.279 | slc3a2a | -0.130 |
| id4 | 0.279 | spon1b | -0.129 |
| sypb | 0.277 | gpr37l1b | -0.126 |
| syn2a | 0.276 | her15.1 | -0.125 |
| zgc:153426 | 0.275 | cldn5a | -0.125 |