Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory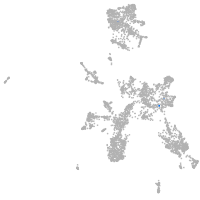 |
otic / lateral line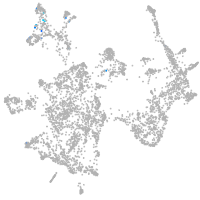 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mss51 | 0.484 | hmgb1b | -0.069 |
| si:ch211-131k2.3 | 0.466 | marcksl1a | -0.065 |
| tnni2b.1 | 0.447 | chd4a | -0.059 |
| tmx2a | 0.441 | si:ch211-288g17.3 | -0.054 |
| smyd1b | 0.437 | stmn1a | -0.054 |
| txlnba | 0.437 | tuba1c | -0.054 |
| txlnbb | 0.429 | cbx5 | -0.052 |
| hsp90aa1.1 | 0.426 | nova2 | -0.052 |
| hjv | 0.425 | tuba1a | -0.050 |
| rbfox1l | 0.423 | rbbp4 | -0.049 |
| zgc:92518 | 0.423 | si:ch211-137a8.4 | -0.049 |
| CR381686.5 | 0.422 | gpm6aa | -0.048 |
| klhl40b | 0.407 | mdkb | -0.048 |
| unc45b | 0.406 | pcna | -0.048 |
| fbxl22 | 0.402 | pfn2 | -0.048 |
| klhl41b | 0.400 | lasp1 | -0.047 |
| zgc:92429 | 0.399 | fabp7a | -0.046 |
| zgc:101853 | 0.395 | gapdhs | -0.046 |
| chrng | 0.389 | mif | -0.046 |
| tnnt2c | 0.387 | hmgn3 | -0.045 |
| myog | 0.386 | prdx2 | -0.045 |
| srl | 0.383 | zc4h2 | -0.045 |
| actc1a | 0.379 | her15.1 | -0.045 |
| obsl1a | 0.375 | foxp4 | -0.044 |
| desma | 0.374 | pdia3 | -0.044 |
| asb12b | 0.369 | si:ch211-156b7.4 | -0.044 |
| klhl31 | 0.362 | ckbb | -0.043 |
| cacng1a | 0.360 | ctsla | -0.043 |
| CABZ01072309.2 | 0.359 | elavl3 | -0.043 |
| chrnd | 0.358 | ywhaqb | -0.043 |
| trim109 | 0.356 | mcm7 | -0.042 |
| acta1a | 0.355 | morf4l1 | -0.042 |
| fitm1 | 0.355 | si:dkey-276j7.1 | -0.042 |
| flncb | 0.355 | sod1 | -0.042 |
| dhrs7ca | 0.351 | tubb5 | -0.042 |




