Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 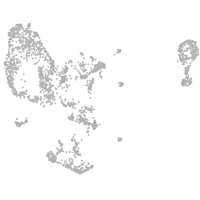 |
neural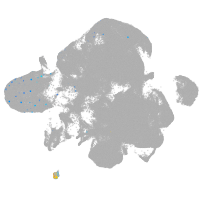 |
spinal cord / glia |
eye |
taste / olfactory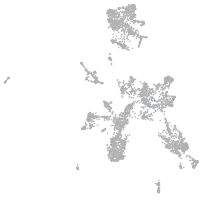 |
otic / lateral line |
epidermis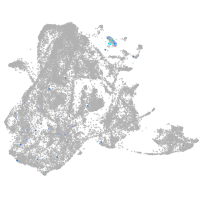 |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01073083.1 | 0.250 | gpm6aa | -0.029 |
| grp | 0.233 | tuba1c | -0.028 |
| si:ch211-256e16.6 | 0.233 | ckbb | -0.027 |
| gfral | 0.218 | stmn1b | -0.026 |
| CR759836.1 | 0.205 | gpm6ab | -0.025 |
| zgc:112408 | 0.160 | gng3 | -0.024 |
| si:ch211-130m23.2 | 0.134 | rtn1a | -0.024 |
| vill | 0.133 | elavl3 | -0.023 |
| itpr3 | 0.132 | cspg5a | -0.022 |
| prr15la | 0.125 | nova2 | -0.022 |
| pvalb8 | 0.115 | hbae3 | -0.021 |
| hepacam2 | 0.111 | hbbe1.3 | -0.021 |
| gcnt7 | 0.110 | rtn1b | -0.021 |
| si:ch211-149l1.2 | 0.110 | ywhag2 | -0.021 |
| pou2f3 | 0.106 | celf2 | -0.020 |
| ponzr5 | 0.104 | elavl4 | -0.020 |
| cldnh | 0.102 | si:dkeyp-75h12.5 | -0.020 |
| si:dkey-188i13.6 | 0.097 | stmn2a | -0.020 |
| zgc:101744 | 0.091 | stx1b | -0.020 |
| zgc:123295 | 0.090 | marcksl1a | -0.020 |
| tyrobp | 0.088 | atpv0e2 | -0.019 |
| npy2r | 0.088 | fabp3 | -0.019 |
| dmrta2 | 0.086 | fez1 | -0.019 |
| tph1b | 0.084 | mdkb | -0.019 |
| itpkca | 0.081 | olfm1b | -0.019 |
| si:ch211-202h22.8 | 0.081 | snap25a | -0.019 |
| LO017877.2 | 0.078 | snap25b | -0.019 |
| penkb | 0.077 | sncb | -0.019 |
| bik | 0.076 | tmsb | -0.019 |
| otofa | 0.076 | tuba1a | -0.019 |
| ovol1a | 0.076 | dpysl2b | -0.019 |
| cers3b | 0.073 | atp6v0cb | -0.018 |
| plcd1a | 0.073 | cadm3 | -0.018 |
| tph1a | 0.073 | fam168a | -0.018 |
| zgc:92380 | 0.070 | gnao1a | -0.018 |




