Other cell groups


all cells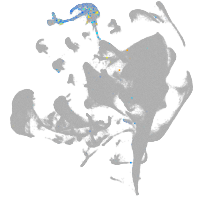 |
blastomeres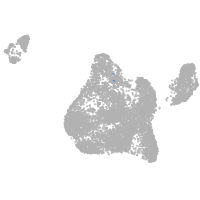 |
PGCs |
endoderm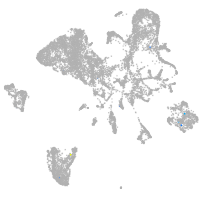 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| p2ry13 | 0.788 | tmsb1 | -0.032 |
| LOC101885690 | 0.788 | s100u | -0.031 |
| itga2b | 0.788 | tmed2 | -0.028 |
| XLOC-016965 | 0.777 | h1f0 | -0.028 |
| AL954142.1 | 0.758 | dad1 | -0.027 |
| mir223 | 0.758 | vps29 | -0.027 |
| susd1 | 0.732 | krt97 | -0.025 |
| fgl2a | 0.700 | hsbp1b | -0.025 |
| LOC100536469 | 0.674 | krt91 | -0.025 |
| plac8l1 | 0.655 | hif1al | -0.025 |
| cd44b | 0.637 | tmed4 | -0.025 |
| gp1bb | 0.636 | blcap | -0.025 |
| BX510919.1 | 0.614 | vma21 | -0.025 |
| grinaa | 0.596 | zgc:92380 | -0.025 |
| si:dkey-85k15.4 | 0.580 | prelid1a | -0.024 |
| prkacba | 0.580 | syap1 | -0.024 |
| plek | 0.559 | sept8a | -0.024 |
| diras2 | 0.554 | tegt | -0.023 |
| btk | 0.543 | gnpnat1 | -0.023 |
| tubb1 | 0.534 | ufm1 | -0.023 |
| rassf2b | 0.525 | lamtor2 | -0.023 |
| fli1a | 0.523 | cox6c | -0.023 |
| fermt3b | 0.500 | prkcsh | -0.023 |
| LOC110439838 | 0.480 | lsm7 | -0.022 |
| DIPK2B | 0.468 | ube2nb | -0.022 |
| CABZ01066694.1 | 0.428 | hspa5 | -0.022 |
| FQ311924.1 | 0.426 | anxa11b | -0.022 |
| si:ch211-214p13.9 | 0.418 | arf5 | -0.022 |
| zgc:113363 | 0.410 | ssr3 | -0.022 |
| BX000363.1 | 0.401 | hibadhb | -0.022 |
| hmha1a | 0.401 | serinc2 | -0.022 |
| myh11a | 0.369 | mrps34 | -0.021 |
| hdr | 0.366 | nupr1b | -0.021 |
| myocd | 0.365 | pmm2 | -0.021 |
| rhd | 0.361 | agr2 | -0.021 |