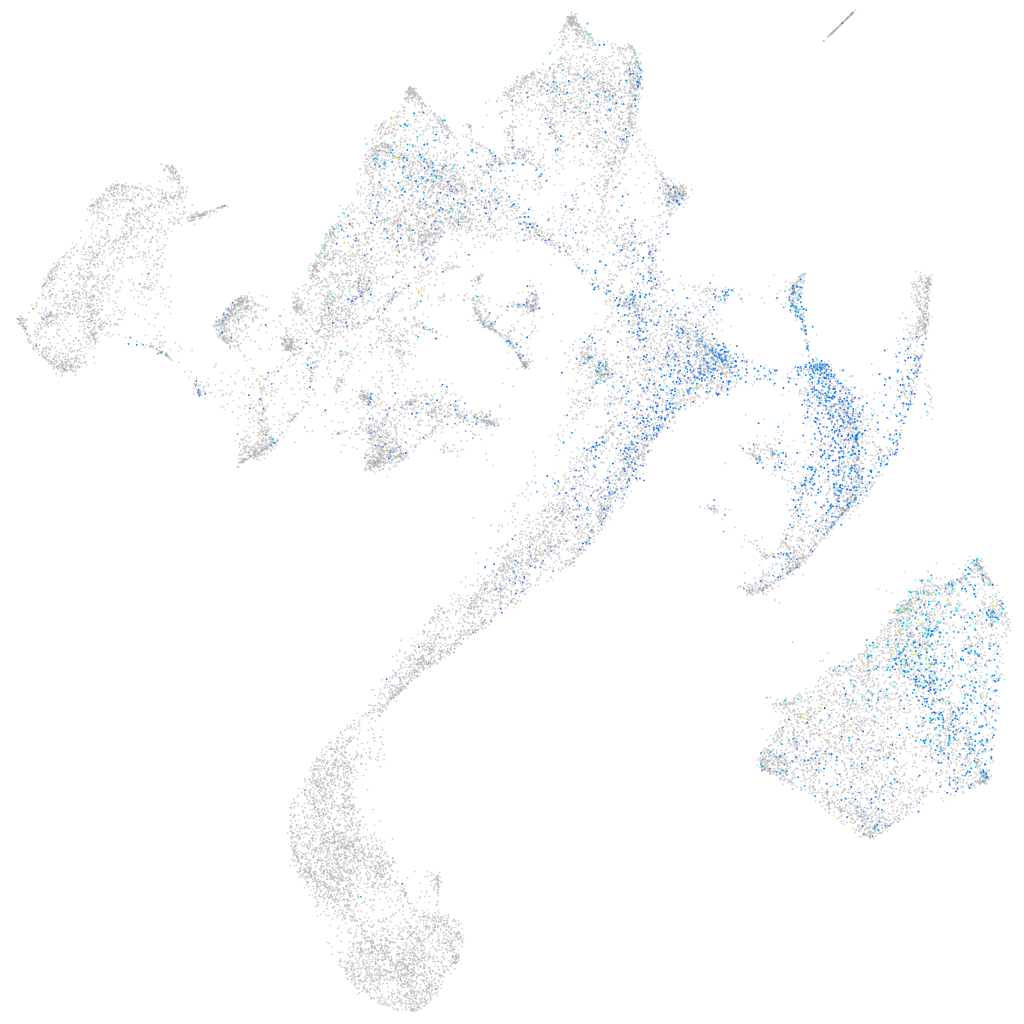
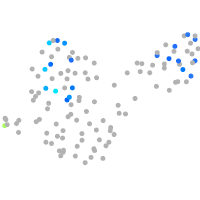
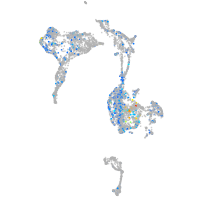
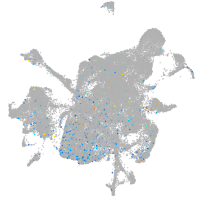
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| efnb2b | 0.208 | actc1b | -0.177 |
| msgn1 | 0.206 | ckmb | -0.160 |
| si:ch73-281n10.2 | 0.192 | ak1 | -0.159 |
| asph | 0.189 | ckma | -0.158 |
| cx43.4 | 0.185 | atp2a1 | -0.156 |
| apoc1 | 0.184 | aldoab | -0.155 |
| tbx6 | 0.183 | tmem38a | -0.153 |
| myf5 | 0.183 | tnnc2 | -0.153 |
| fn1b | 0.181 | acta1b | -0.150 |
| nid2a | 0.177 | mylpfa | -0.149 |
| syncrip | 0.176 | neb | -0.147 |
| hmgb2a | 0.175 | eno1a | -0.146 |
| hmga1a | 0.173 | tpma | -0.144 |
| dlc | 0.170 | pabpc4 | -0.144 |
| pabpc1a | 0.169 | ldb3b | -0.141 |
| nop58 | 0.169 | gapdh | -0.141 |
| tbx16 | 0.168 | ttn.2 | -0.141 |
| tbx16l | 0.166 | CABZ01078594.1 | -0.140 |
| ncl | 0.166 | mylz3 | -0.139 |
| khdrbs1a | 0.165 | ldb3a | -0.139 |
| pcdh8 | 0.165 | pvalb2 | -0.139 |
| hes6 | 0.164 | pvalb1 | -0.138 |
| apoeb | 0.164 | tpi1b | -0.138 |
| h2afvb | 0.163 | si:ch73-367p23.2 | -0.138 |
| nop56 | 0.163 | tnnt3a | -0.138 |
| anp32b | 0.162 | actn3a | -0.138 |
| hnrnpaba | 0.162 | eno3 | -0.136 |
| im:7138239 | 0.161 | cav3 | -0.136 |
| aplnra | 0.161 | mybphb | -0.136 |
| dld | 0.160 | gamt | -0.136 |
| npm1a | 0.160 | srl | -0.136 |
| gnaia | 0.160 | desma | -0.136 |
| si:ch73-1a9.3 | 0.159 | actn3b | -0.135 |
| hmgb2b | 0.159 | smyd1a | -0.134 |
| cirbpa | 0.158 | tmod4 | -0.133 |