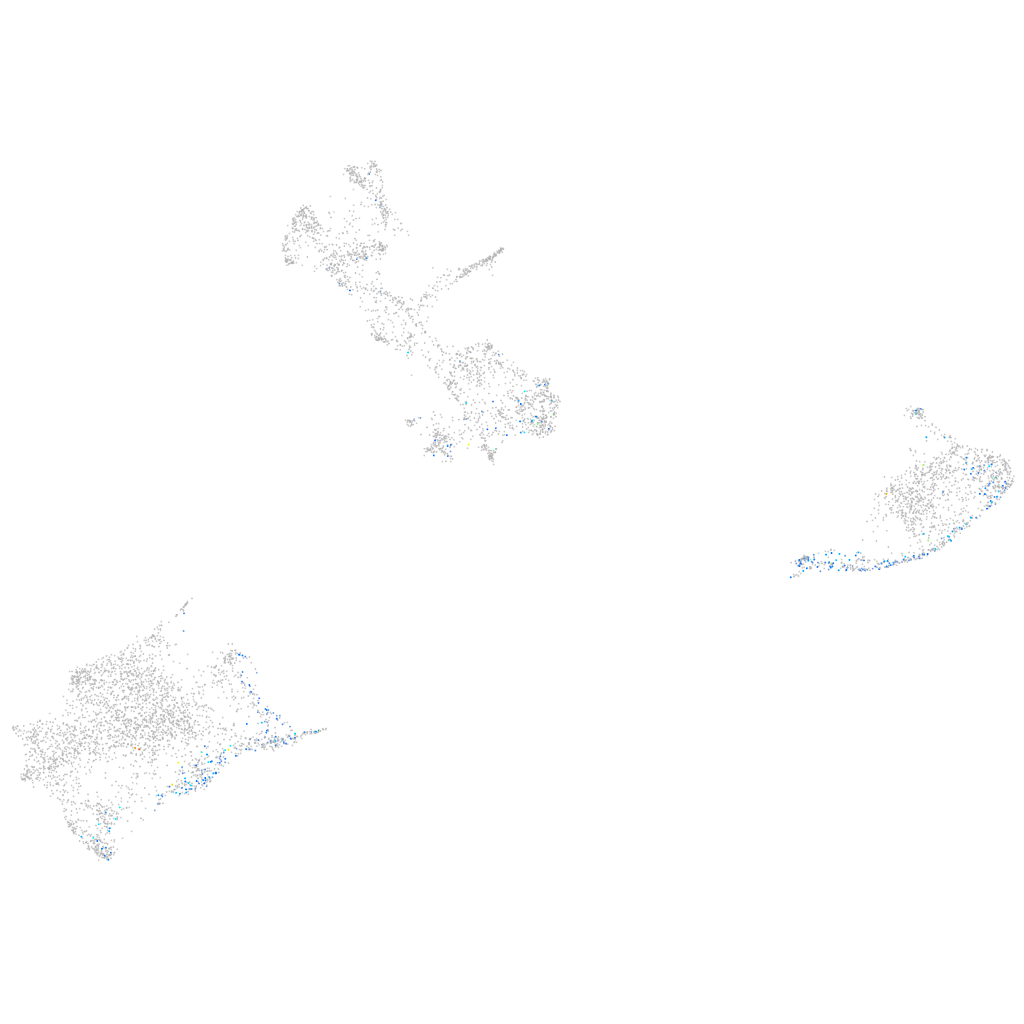
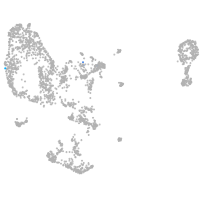
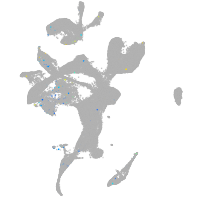
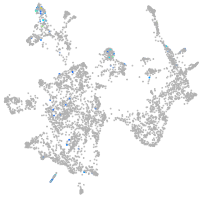
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pim1 | 0.206 | pvalb2 | -0.094 |
| pcdh9 | 0.199 | uraha | -0.078 |
| crestin | 0.172 | defbl1 | -0.077 |
| xbp1 | 0.163 | apoda.1 | -0.076 |
| mitfa | 0.162 | gpnmb | -0.071 |
| canx | 0.160 | si:ch211-256m1.8 | -0.071 |
| SPAG9 | 0.158 | actc1b | -0.070 |
| ddit3 | 0.157 | pvalb1 | -0.069 |
| eva1ba | 0.156 | si:dkey-197i20.6 | -0.065 |
| tfap2e | 0.154 | phyhd1 | -0.064 |
| phlda1 | 0.153 | zgc:110339 | -0.064 |
| CR383676.1 | 0.152 | lypc | -0.064 |
| ctsc | 0.151 | tmsb4x | -0.063 |
| msx1b | 0.148 | si:ch73-89b15.3 | -0.062 |
| atp6ap1b | 0.147 | alx4a | -0.061 |
| psap | 0.143 | s100v2 | -0.060 |
| trpm1b | 0.141 | mylpfa | -0.057 |
| XLOC-034167 | 0.139 | sytl2b | -0.057 |
| si:ch211-122f10.4 | 0.138 | hbae3 | -0.056 |
| atf3 | 0.134 | krt18b | -0.056 |
| ctsba | 0.134 | myhz1.1 | -0.055 |
| tram1 | 0.133 | akap12a | -0.055 |
| itga4 | 0.132 | rpl12 | -0.055 |
| tfap2a | 0.131 | tnni2a.4 | -0.053 |
| si:dkeyp-110e4.6 | 0.131 | alx4b | -0.052 |
| zeb2a | 0.131 | cdh11 | -0.052 |
| mcf2l2 | 0.130 | unm-sa821 | -0.051 |
| cdh1 | 0.129 | alx1 | -0.051 |
| naga | 0.129 | prx | -0.050 |
| znf1000 | 0.129 | ponzr1 | -0.050 |
| lman2 | 0.129 | si:ch211-105c13.3 | -0.050 |
| LOC110438542 | 0.129 | myof | -0.050 |
| atp6ap2 | 0.129 | mylz3 | -0.049 |
| slc3a2a | 0.128 | guk1a | -0.048 |
| hspd1 | 0.128 | tmem179b | -0.048 |