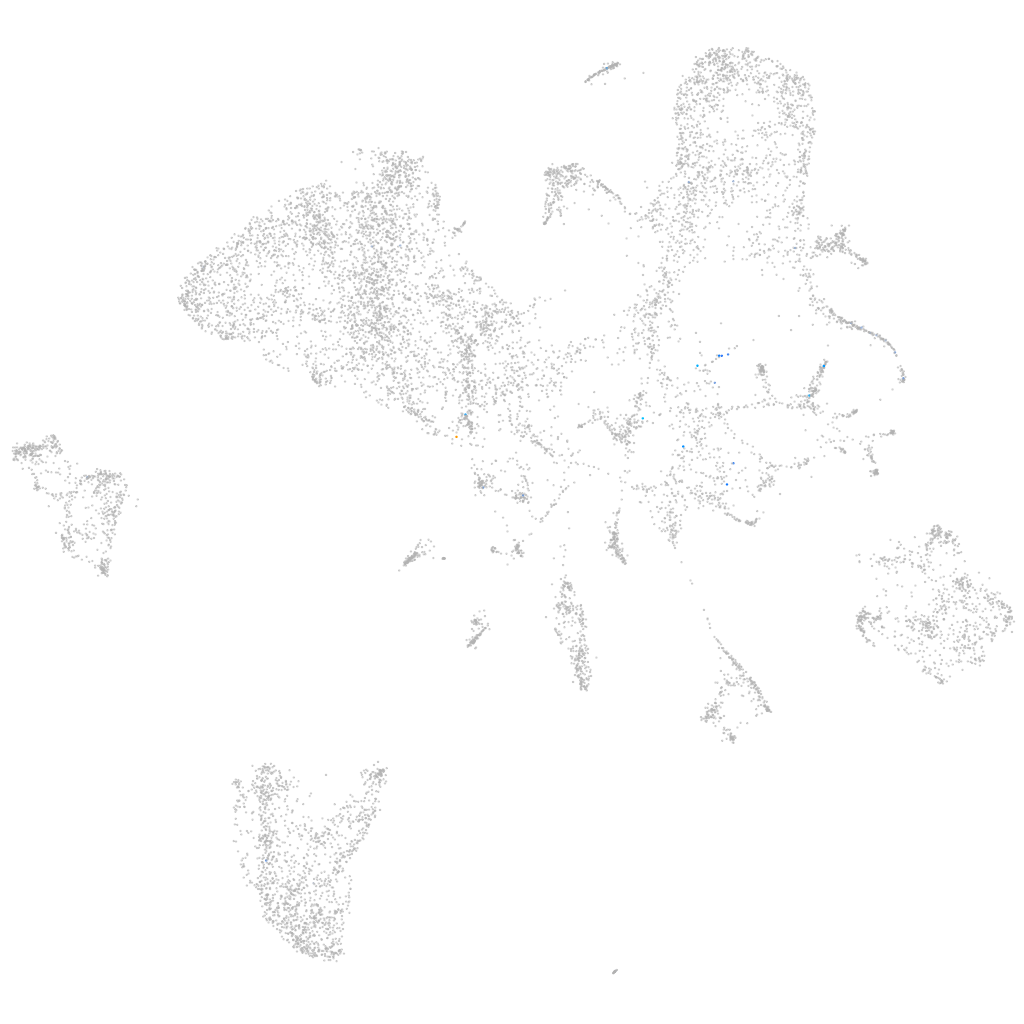
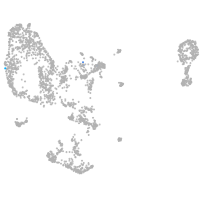
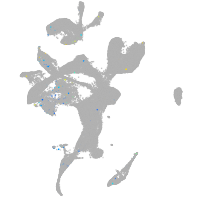
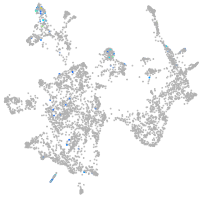
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gabrz | 0.580 | dap | -0.036 |
| XLOC-025875 | 0.464 | gamt | -0.036 |
| XLOC-002533 | 0.436 | ssr2 | -0.035 |
| nlrp16 | 0.395 | nupr1b | -0.034 |
| gfra2b | 0.382 | ddt | -0.034 |
| XLOC-005257 | 0.374 | slc25a5 | -0.033 |
| gabrd | 0.327 | rplp1 | -0.033 |
| apba2a | 0.320 | cx32.3 | -0.032 |
| si:ch211-288d18.1 | 0.311 | bhmt | -0.031 |
| CU024870.1 | 0.311 | scp2a | -0.031 |
| admb | 0.306 | fbp1b | -0.030 |
| pcdh2ab11 | 0.306 | glud1b | -0.030 |
| pcdh1g32 | 0.305 | sec61g | -0.030 |
| kcnd3 | 0.303 | ssr4 | -0.030 |
| ccsapa | 0.294 | mat1a | -0.030 |
| LOC100536761 | 0.289 | apoa4b.1 | -0.029 |
| gabra5 | 0.285 | afp4 | -0.029 |
| BX548075.3 | 0.276 | ahcy | -0.029 |
| ccni2 | 0.273 | cdo1 | -0.029 |
| zgc:152863 | 0.268 | acadm | -0.029 |
| cacnb2a | 0.265 | rps2 | -0.029 |
| syt9a | 0.264 | suclg2 | -0.029 |
| slc12a5a | 0.256 | rps19 | -0.028 |
| fibcd1 | 0.253 | apoc2 | -0.028 |
| si:ch73-290k24.6 | 0.252 | gpx4b | -0.028 |
| kcnn1a | 0.247 | mdh1aa | -0.028 |
| itgb8 | 0.246 | gcshb | -0.027 |
| cdh10a | 0.245 | echs1 | -0.027 |
| myo3a | 0.240 | agxtb | -0.027 |
| fstl5 | 0.239 | gapdh | -0.027 |
| spon1a | 0.237 | mgst1.2 | -0.027 |
| LOC101884792 | 0.237 | hspe1 | -0.027 |
| pip4k2ab | 0.234 | sec61a1 | -0.027 |
| gpr158a | 0.234 | dhrs9 | -0.026 |
| cnih2 | 0.234 | atp5pb | -0.026 |