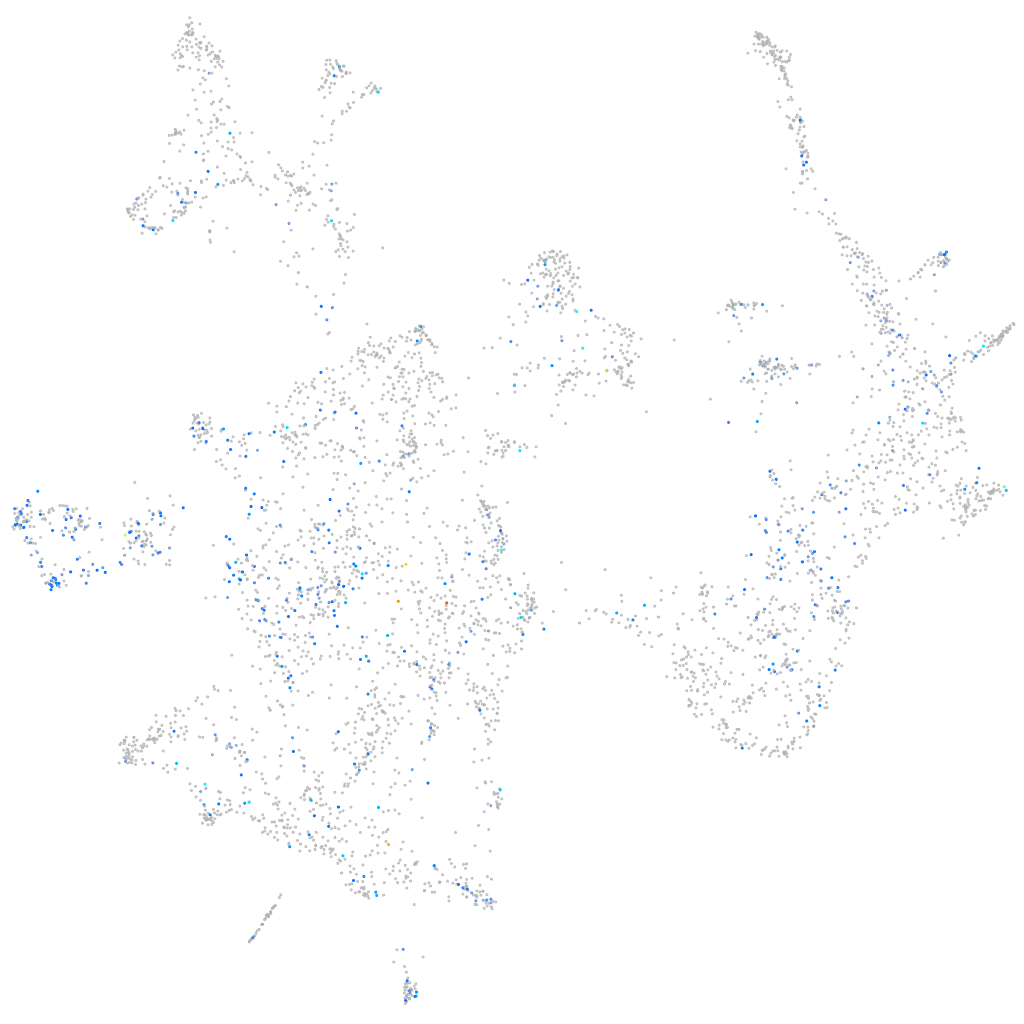
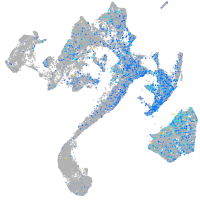
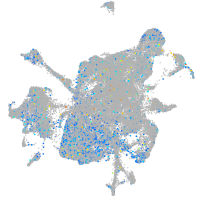
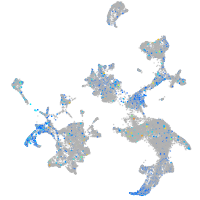
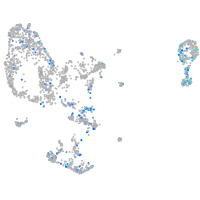
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccna2 | 0.186 | COX3 | -0.095 |
| rpa2 | 0.174 | mt-co2 | -0.076 |
| ranbp1 | 0.166 | rnasekb | -0.072 |
| shmt1 | 0.165 | kif1aa | -0.068 |
| pcna | 0.164 | si:ch73-15n15.3 | -0.067 |
| rrm1 | 0.163 | zgc:158463 | -0.067 |
| fen1 | 0.163 | pvalb8 | -0.067 |
| hmgb2a | 0.161 | calm1b | -0.066 |
| chaf1a | 0.160 | pvalb2 | -0.066 |
| mcm7 | 0.159 | slc17a8 | -0.064 |
| npm1a | 0.157 | nptna | -0.064 |
| dut | 0.155 | CR383676.1 | -0.063 |
| stmn1a | 0.154 | pou4f3 | -0.063 |
| orc4 | 0.151 | dnajc5b | -0.063 |
| anp32b | 0.150 | calm1a | -0.062 |
| cnbpa | 0.150 | ptprga | -0.061 |
| ran | 0.149 | mb | -0.061 |
| ddx39ab | 0.146 | emb | -0.061 |
| zgc:110216 | 0.144 | mt-co1 | -0.060 |
| cdca8 | 0.144 | osbpl1a | -0.060 |
| nasp | 0.144 | CR925719.1 | -0.059 |
| h3f3a | 0.144 | glula | -0.059 |
| mad2l1 | 0.142 | copz2 | -0.059 |
| cdca5 | 0.141 | mir7a-1 | -0.058 |
| mis12 | 0.141 | erich3 | -0.058 |
| cks1b | 0.141 | gngt2b | -0.058 |
| mki67 | 0.139 | gapdhs | -0.058 |
| banf1 | 0.139 | gipc3 | -0.057 |
| cdk1 | 0.139 | zgc:162989 | -0.057 |
| cdca7b | 0.139 | tpi1a | -0.057 |
| msh6 | 0.137 | tex33 | -0.057 |
| ccnb1 | 0.136 | clic5b | -0.057 |
| lig1 | 0.136 | twf2b | -0.057 |
| tubb2b | 0.135 | grtp1a | -0.056 |
| aurkb | 0.135 | slitrk6 | -0.056 |