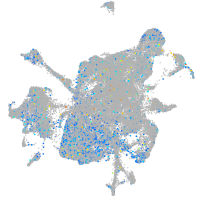
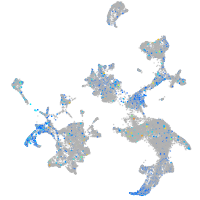
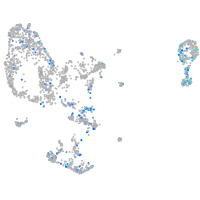
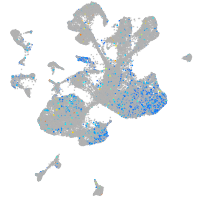
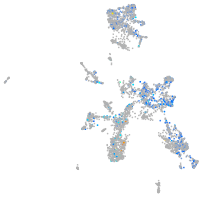
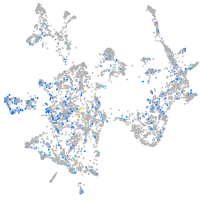
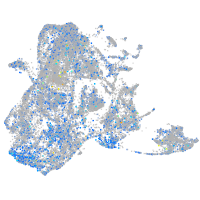
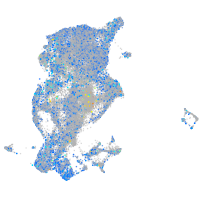
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dut | 0.170 | rtn1a | -0.096 |
| pcna | 0.169 | ckbb | -0.095 |
| npm1a | 0.166 | myt1b | -0.091 |
| rpa2 | 0.163 | elavl3 | -0.090 |
| nutf2l | 0.162 | gpm6aa | -0.085 |
| anp32b | 0.157 | hbbe1.3 | -0.083 |
| cks1b | 0.156 | pvalb1 | -0.083 |
| dkc1 | 0.155 | pvalb2 | -0.083 |
| fen1 | 0.154 | COX3 | -0.082 |
| cdx4 | 0.153 | actc1b | -0.082 |
| chaf1a | 0.153 | CR383676.1 | -0.079 |
| nop58 | 0.152 | rnasekb | -0.076 |
| hspb1 | 0.152 | tmsb | -0.075 |
| ranbp1 | 0.152 | gpm6ab | -0.072 |
| rrm1 | 0.149 | hbae3 | -0.072 |
| nasp | 0.148 | marcksl1b | -0.069 |
| ccna2 | 0.148 | dpysl3 | -0.068 |
| mcm7 | 0.148 | nova2 | -0.068 |
| mki67 | 0.147 | hbae1.1 | -0.066 |
| rpa3 | 0.146 | calm1a | -0.065 |
| hmgb2a | 0.145 | si:dkeyp-75h12.5 | -0.065 |
| selenoh | 0.144 | gapdhs | -0.065 |
| lig1 | 0.143 | stxbp1a | -0.065 |
| dek | 0.143 | dpysl2b | -0.065 |
| rrm2 | 0.142 | stx1b | -0.065 |
| lbr | 0.142 | gnao1a | -0.064 |
| mad2l1 | 0.141 | vamp2 | -0.064 |
| snu13b | 0.141 | atp6v0cb | -0.064 |
| banf1 | 0.141 | cotl1 | -0.063 |
| stmn1a | 0.140 | mylpfa | -0.063 |
| cbx3a | 0.140 | syt2a | -0.063 |
| nhp2 | 0.139 | ywhah | -0.063 |
| rnaseh2a | 0.138 | vat1 | -0.062 |
| tuba8l4 | 0.137 | ube2ql1 | -0.062 |
| shmt1 | 0.137 | cplx2 | -0.061 |