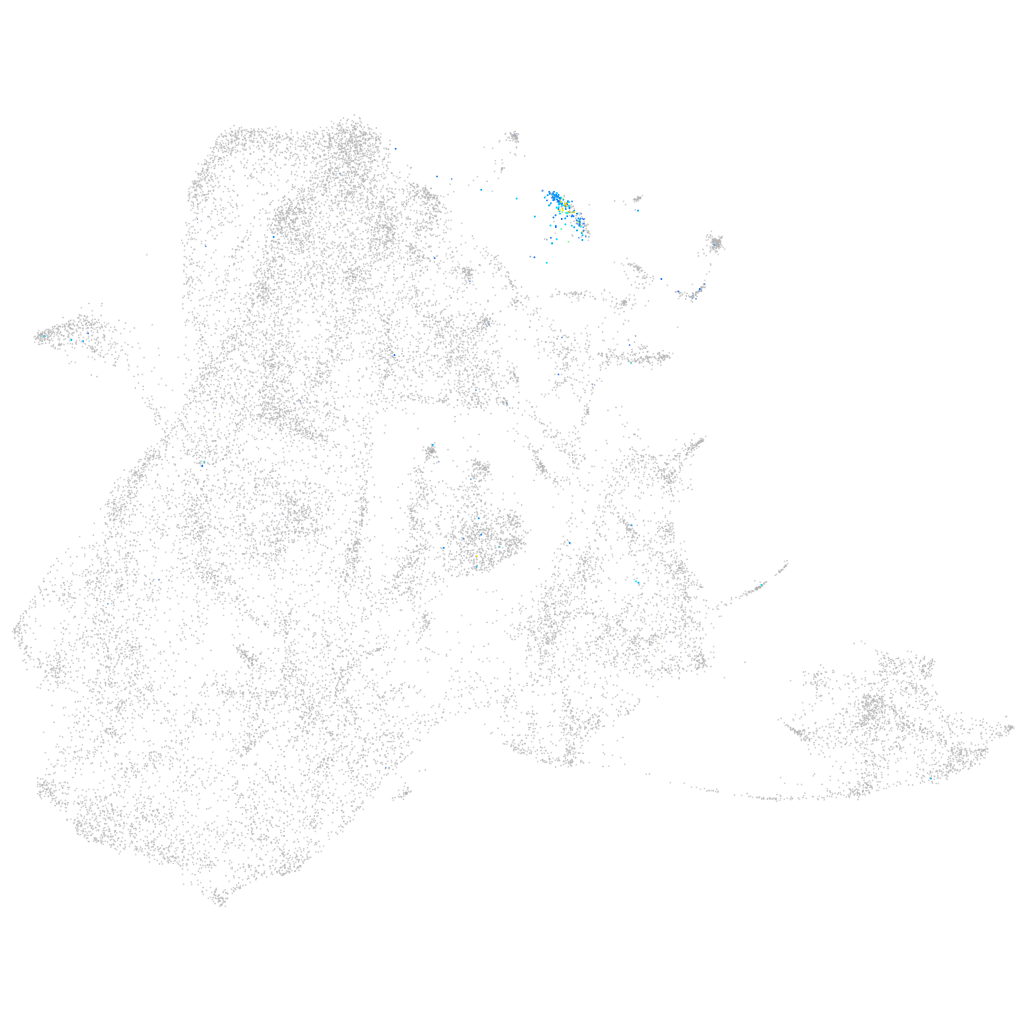
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory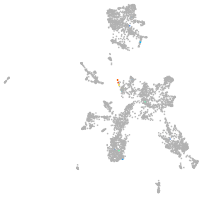 |
otic / lateral line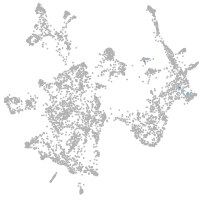 |
epidermis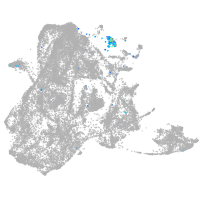 |
periderm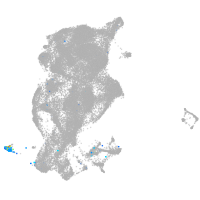 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| grp | 0.677 | rps7 | -0.063 |
| CR759836.1 | 0.615 | rps15a | -0.052 |
| si:ch211-256e16.6 | 0.581 | rpl12 | -0.052 |
| gfral | 0.539 | rps19 | -0.052 |
| pvalb8 | 0.417 | snrpb | -0.051 |
| prr15la | 0.399 | ssr3 | -0.051 |
| vill | 0.397 | rpl23 | -0.051 |
| tyrobp | 0.360 | rplp2l | -0.051 |
| zgc:112408 | 0.356 | khdrbs1a | -0.050 |
| si:ch211-149l1.2 | 0.347 | rpl13a | -0.049 |
| CT583687.1 | 0.342 | ptges3b | -0.049 |
| itpr3 | 0.321 | rps23 | -0.048 |
| myo15aa | 0.299 | rpn1 | -0.048 |
| BX247868.1 | 0.284 | rps26 | -0.048 |
| otofa | 0.270 | rpl19 | -0.048 |
| gcnt7 | 0.263 | rps9 | -0.047 |
| chga | 0.261 | rps4x | -0.047 |
| zgc:123295 | 0.247 | krt18a.1 | -0.047 |
| cldnh | 0.241 | rpl17 | -0.046 |
| cobl | 0.237 | rpl7a | -0.046 |
| penkb | 0.236 | hmga1a | -0.046 |
| dmrta2 | 0.233 | rplp0 | -0.046 |
| pcbp3 | 0.225 | hmgn6 | -0.046 |
| pcsk2 | 0.225 | rps27.1 | -0.045 |
| cngb3.2 | 0.222 | ybx1 | -0.045 |
| hepacam2 | 0.222 | hsp90ab1 | -0.045 |
| si:ch211-256e16.4 | 0.211 | rps20 | -0.045 |
| si:ch211-130m23.2 | 0.209 | syncrip | -0.044 |
| LOC108182217 | 0.206 | apex1 | -0.044 |
| CABZ01095001.1 | 0.205 | faua | -0.044 |
| scg2a | 0.205 | rpl10a | -0.043 |
| LO017877.2 | 0.202 | ssr4 | -0.043 |
| si:dkey-222f2.1 | 0.198 | ilf2 | -0.043 |
| cers3b | 0.190 | rpl9 | -0.043 |
| npy2r | 0.189 | sub1a | -0.043 |