"2,3-bisphosphoglycerate mutase"
ZFIN 
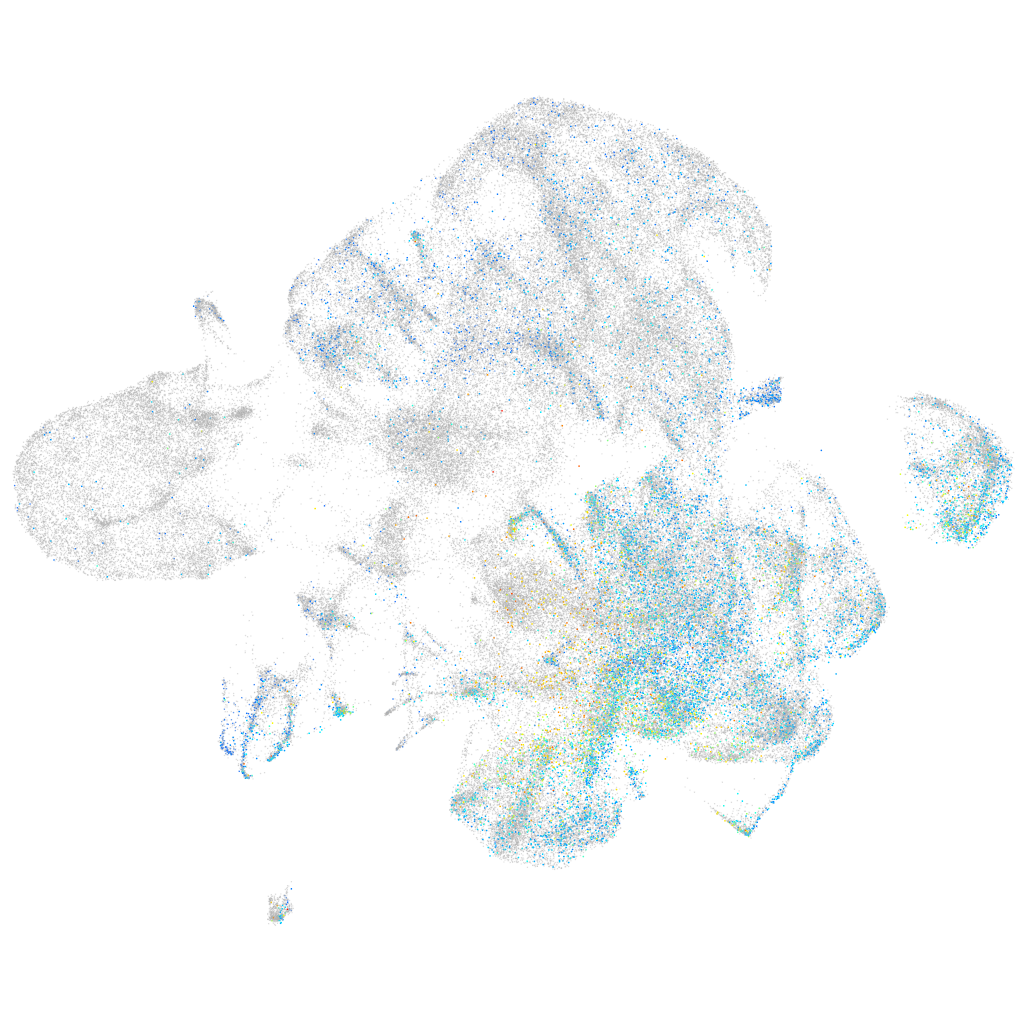
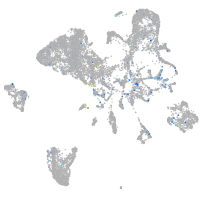
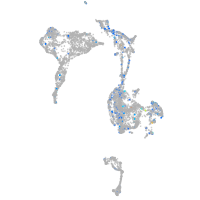
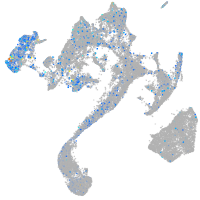
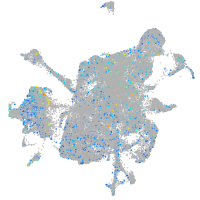
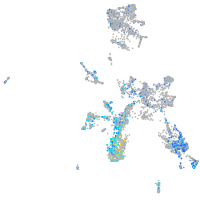
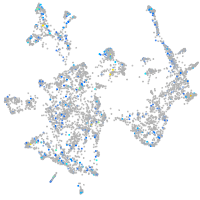
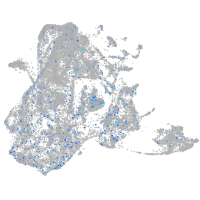
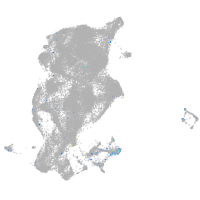
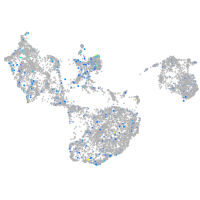
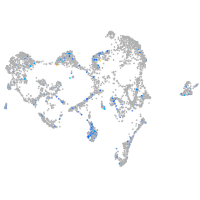
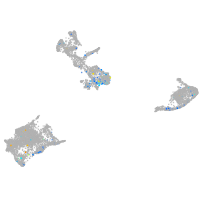
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ywhag2 | 0.221 | hmgb2a | -0.176 |
| sncb | 0.220 | hmga1a | -0.157 |
| gapdhs | 0.216 | hmgb2b | -0.144 |
| atp6v1e1b | 0.215 | si:dkey-151g10.6 | -0.143 |
| zgc:65894 | 0.215 | rplp2l | -0.135 |
| gng3 | 0.213 | id1 | -0.134 |
| atp6v0cb | 0.211 | rps12 | -0.131 |
| atp6v1g1 | 0.210 | rplp1 | -0.131 |
| stmn2a | 0.206 | pcna | -0.129 |
| snap25a | 0.206 | npm1a | -0.124 |
| vamp2 | 0.205 | stmn1a | -0.124 |
| stxbp1a | 0.199 | rps28 | -0.123 |
| tpi1b | 0.197 | nop58 | -0.121 |
| calm1a | 0.194 | eef1a1l1 | -0.121 |
| mllt11 | 0.194 | ranbp1 | -0.118 |
| rnasekb | 0.193 | snu13b | -0.117 |
| stx1b | 0.191 | rpl29 | -0.116 |
| tmsb2 | 0.191 | dkc1 | -0.115 |
| elavl4 | 0.188 | nop56 | -0.114 |
| calm1b | 0.187 | rps20 | -0.113 |
| ckbb | 0.186 | mdka | -0.112 |
| eno1a | 0.185 | chaf1a | -0.112 |
| calm2a | 0.185 | anp32b | -0.111 |
| map1aa | 0.185 | ccnd1 | -0.110 |
| stmn1b | 0.184 | XLOC-003690 | -0.110 |
| sncgb | 0.183 | mcm6 | -0.110 |
| gap43 | 0.182 | msna | -0.110 |
| syn2a | 0.182 | sox19a | -0.110 |
| atpv0e2 | 0.180 | mcm7 | -0.109 |
| rtn1b | 0.178 | fbl | -0.109 |
| tuba1c | 0.178 | sox3 | -0.109 |
| rab6bb | 0.177 | cx43.4 | -0.109 |
| eno2 | 0.177 | snrpb | -0.108 |
| gpm6ab | 0.176 | rpl4 | -0.108 |
| atp6ap2 | 0.176 | rpsa | -0.108 |