bone morphogenetic protein 2b
ZFIN 
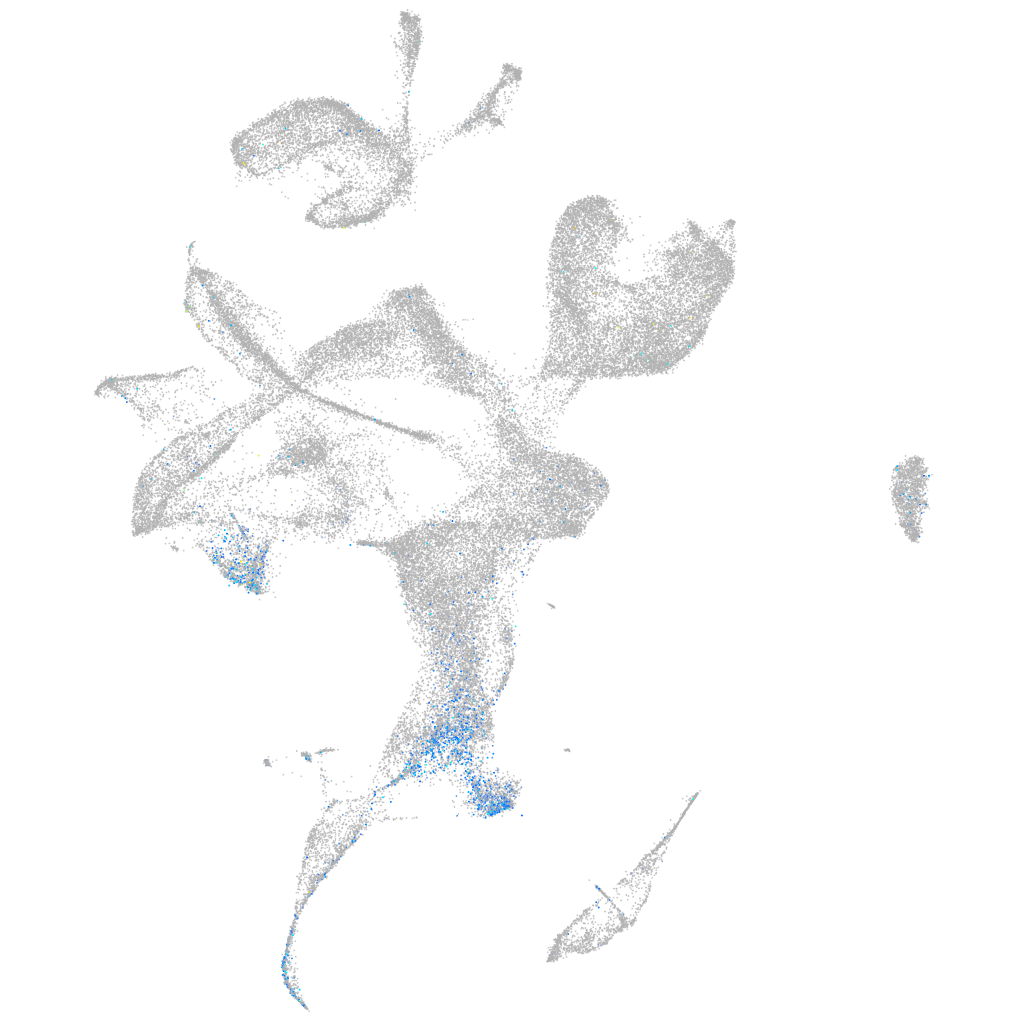
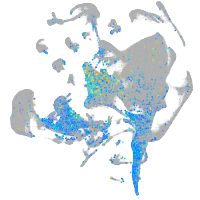
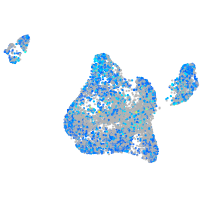
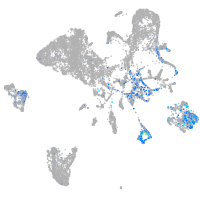
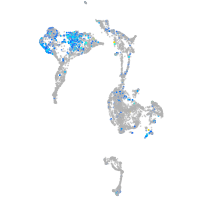
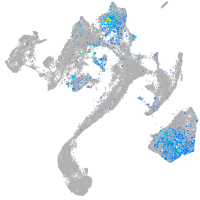
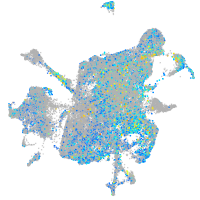
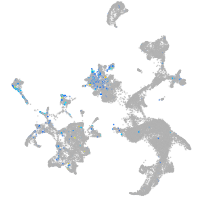
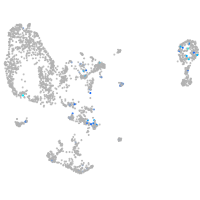
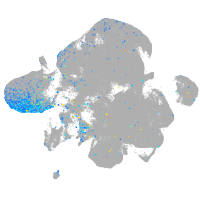
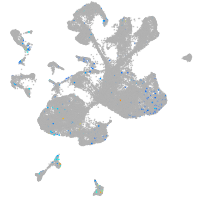
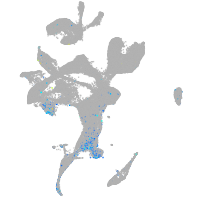
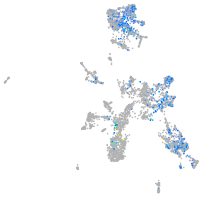
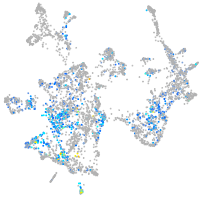
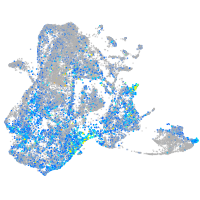
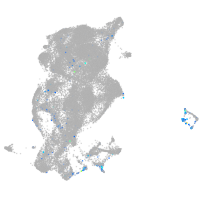
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| aldh1a2 | 0.239 | ptmab | -0.126 |
| bmp7b | 0.173 | hmgn6 | -0.103 |
| zfp36l1a | 0.172 | gpm6aa | -0.093 |
| slc38a5b | 0.167 | ckbb | -0.091 |
| cdon | 0.165 | si:ch73-1a9.3 | -0.081 |
| cdkn1a | 0.160 | tuba1c | -0.079 |
| bambia | 0.158 | gpm6ab | -0.077 |
| eif4ebp3l | 0.158 | hmgb1a | -0.073 |
| CU929070.1 | 0.155 | ptmaa | -0.069 |
| npm1a | 0.154 | cadm3 | -0.069 |
| si:dkey-102m7.3 | 0.153 | btg1 | -0.067 |
| paics | 0.153 | epb41a | -0.067 |
| tbx5a | 0.151 | neurod4 | -0.067 |
| fosab | 0.149 | marcksl1b | -0.066 |
| adi1 | 0.148 | scrt2 | -0.066 |
| rcl1 | 0.148 | ndrg4 | -0.066 |
| id1 | 0.147 | snap25b | -0.065 |
| atic | 0.147 | pvalb1 | -0.064 |
| cad | 0.146 | atp6v0cb | -0.064 |
| prps1a | 0.142 | rnasekb | -0.064 |
| sparc | 0.141 | rtn1a | -0.062 |
| mfap2 | 0.141 | foxg1b | -0.062 |
| nhp2 | 0.138 | crx | -0.062 |
| snu13b | 0.138 | tuba1a | -0.061 |
| rps27l | 0.136 | mpp6b | -0.061 |
| nop58 | 0.134 | rorb | -0.060 |
| jun | 0.134 | stmn1b | -0.060 |
| si:dkey-7j14.6 | 0.134 | pvalb2 | -0.059 |
| gart | 0.133 | h3f3c | -0.059 |
| aldob | 0.132 | hbbe1.3 | -0.058 |
| dkc1 | 0.132 | tp53inp2 | -0.057 |
| nip7 | 0.131 | ubc | -0.056 |
| rsl1d1 | 0.130 | anp32e | -0.056 |
| gstm.1 | 0.130 | atp1b2a | -0.056 |
| nop10 | 0.128 | syt5b | -0.056 |