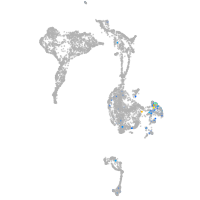
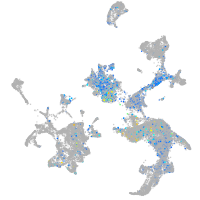
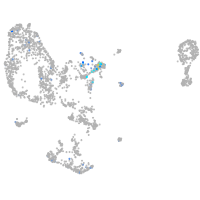
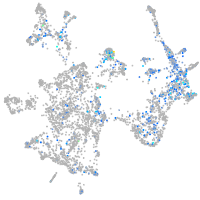
BAF chromatin remodeling complex subunit BCL11A a
ZFIN 
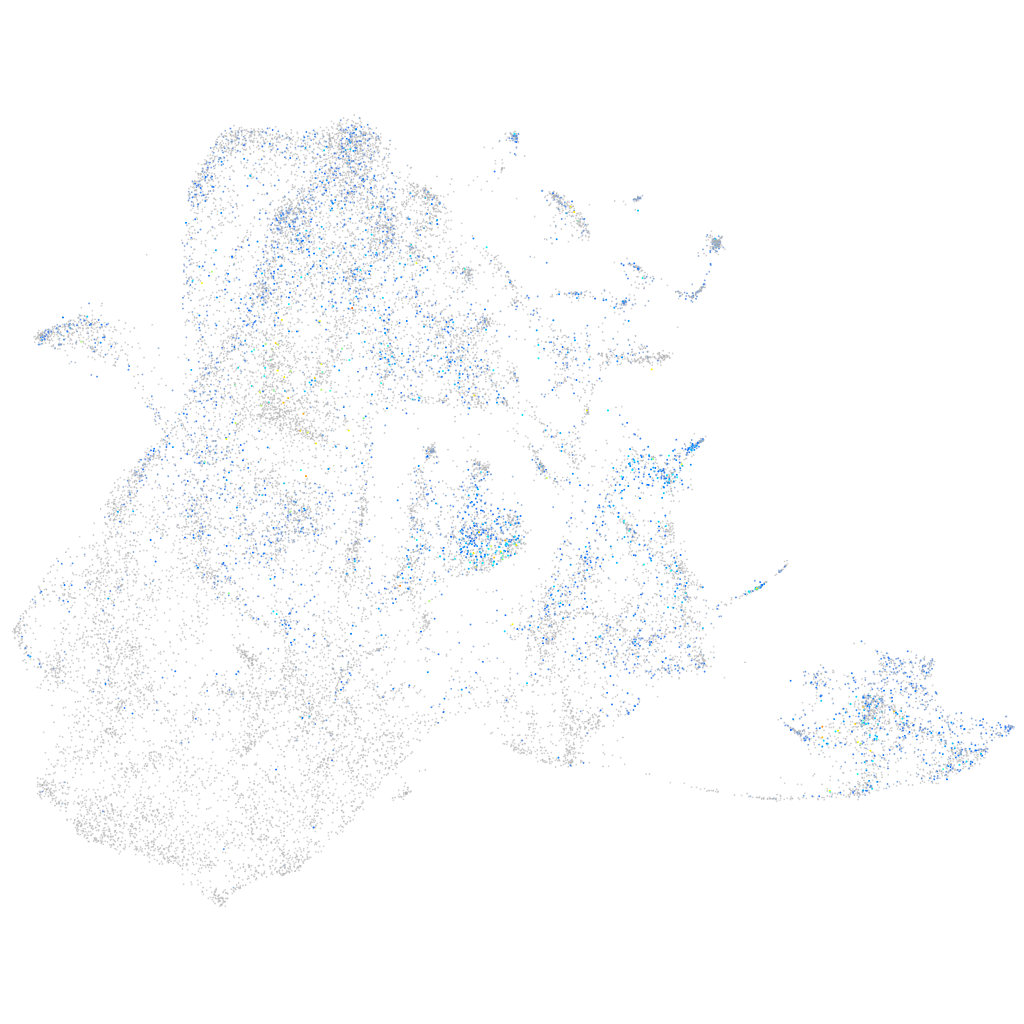
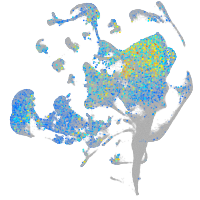
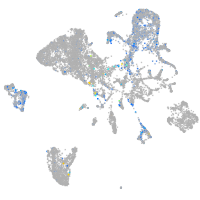
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rhbg | 0.134 | lin28a | -0.136 |
| COX3 | 0.128 | nid2a | -0.134 |
| gpm6aa | 0.126 | tpm4a | -0.132 |
| gpm6ab | 0.125 | CABZ01075068.1 | -0.129 |
| syt1a | 0.124 | myl6 | -0.120 |
| sod1 | 0.123 | BX927258.1 | -0.120 |
| rplp2 | 0.120 | hspb1 | -0.118 |
| bcl11ba | 0.119 | sdr16c5a | -0.113 |
| olfm1b | 0.118 | apoc1 | -0.110 |
| mt-co2 | 0.117 | akap12b | -0.109 |
| stmn1b | 0.116 | mdka | -0.108 |
| nme2b.1 | 0.116 | XLOC-032526 | -0.107 |
| sncb | 0.115 | XLOC-001964 | -0.107 |
| pbx3b | 0.115 | ppp1r3b | -0.105 |
| ppl | 0.114 | cx43.4 | -0.105 |
| egr1 | 0.114 | fbn2a.1 | -0.105 |
| tuba1c | 0.112 | odc1 | -0.103 |
| rnasekb | 0.111 | krt8 | -0.103 |
| eef1da | 0.110 | p3h1 | -0.102 |
| snap25a | 0.110 | ssr3 | -0.100 |
| mt-atp6 | 0.110 | cnn2 | -0.100 |
| crip1 | 0.110 | crtap | -0.100 |
| ndrg2 | 0.109 | fn1a | -0.099 |
| gapdhs | 0.109 | rrbp1b | -0.099 |
| atp6v0cb | 0.109 | ssr4 | -0.099 |
| gng3 | 0.108 | fbn2b | -0.098 |
| gcm2 | 0.108 | trim71 | -0.098 |
| evplb | 0.108 | gpx8 | -0.094 |
| kdm6bb | 0.108 | akt1s1 | -0.093 |
| rtn1b | 0.107 | tspan7 | -0.092 |
| elavl4 | 0.107 | si:dkey-12j5.1 | -0.092 |
| elavl3 | 0.107 | arid3b | -0.091 |
| ociad2 | 0.106 | XLOC-039121 | -0.091 |
| nova2 | 0.105 | tram1 | -0.091 |
| pvalb1 | 0.104 | fkbp10b | -0.091 |