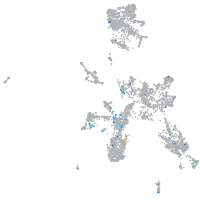
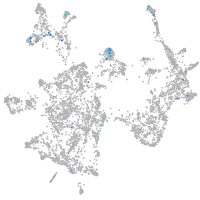
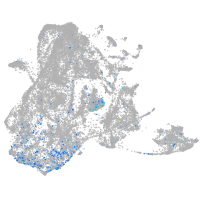
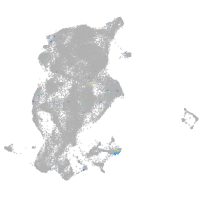
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| camta1a | 0.216 | rpl6 | -0.037 |
| rtn1b | 0.209 | hspb1 | -0.037 |
| stmn2a | 0.205 | dkc1 | -0.035 |
| gpm6aa | 0.203 | tuba8l2 | -0.035 |
| sncb | 0.202 | hsp90aa1.1 | -0.034 |
| LOC103911689 | 0.201 | rrs1 | -0.032 |
| snap25a | 0.195 | rpl4 | -0.030 |
| stmn1b | 0.194 | npm1a | -0.030 |
| stox2a | 0.193 | plp2 | -0.029 |
| mir132-2 | 0.189 | apoc1 | -0.029 |
| vamp2 | 0.189 | six4a | -0.029 |
| aplp1 | 0.189 | UTP14C | -0.029 |
| gng3 | 0.184 | fbl | -0.029 |
| gabrg2 | 0.183 | nop58 | -0.029 |
| cdk5r1b | 0.179 | ubap2b | -0.028 |
| sncgb | 0.179 | ddx18 | -0.028 |
| stxbp1a | 0.179 | rrp1 | -0.028 |
| zgc:153426 | 0.178 | nop56 | -0.028 |
| sypa | 0.178 | tsr1 | -0.028 |
| stx1b | 0.178 | NC-002333.4 | -0.028 |
| vsnl1b | 0.177 | ebna1bp2 | -0.028 |
| eno2 | 0.177 | zgc:92429 | -0.027 |
| elavl4 | 0.176 | BX276101.1 | -0.027 |
| si:ch73-127m5.1 | 0.176 | ntmt1 | -0.027 |
| zgc:65894 | 0.175 | eif4bb | -0.027 |
| elavl3 | 0.174 | eif4ebp3l | -0.027 |
| spock2 | 0.173 | hacd1 | -0.027 |
| kcnc3a | 0.173 | rps18 | -0.027 |
| gap43 | 0.173 | banf1 | -0.027 |
| gnao1a | 0.171 | cdx4 | -0.027 |
| st8sia5 | 0.169 | nop2 | -0.027 |
| maptb | 0.169 | txlnbb | -0.026 |
| mapk8ip2 | 0.168 | tbx16 | -0.026 |
| cplx2l | 0.168 | cdx1a | -0.026 |
| prrt1 | 0.166 | rsl1d1 | -0.026 |