ATRX chromatin remodeler
ZFIN 
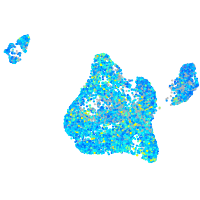
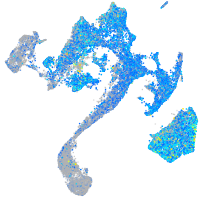
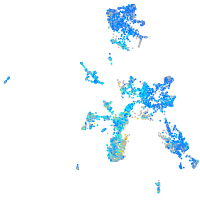
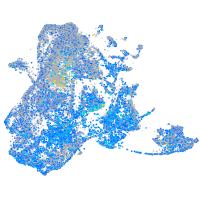
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgb1b | 0.270 | suclg1 | -0.193 |
| tubb2b | 0.259 | gpd1b | -0.190 |
| tmeff1b | 0.252 | aldob | -0.188 |
| khdrbs1a | 0.249 | dab2 | -0.185 |
| ewsr1a | 0.249 | pklr | -0.180 |
| chd4a | 0.247 | eno3 | -0.175 |
| si:ch211-288g17.3 | 0.246 | zgc:136493 | -0.174 |
| elavl3 | 0.245 | si:ch211-139a5.9 | -0.171 |
| hmgb3a | 0.244 | pdzk1 | -0.171 |
| rtn1a | 0.244 | gapdh | -0.170 |
| marcksl1b | 0.242 | zgc:101040 | -0.169 |
| FO082781.1 | 0.238 | nit2 | -0.167 |
| hnrnpaba | 0.237 | si:dkey-194e6.1 | -0.166 |
| h2afvb | 0.236 | grhprb | -0.166 |
| ptmab | 0.234 | fbp1b | -0.164 |
| hmgb1a | 0.233 | dera | -0.164 |
| hmgn2 | 0.232 | pnp6 | -0.164 |
| si:dkey-276j7.1 | 0.231 | atp5mc3b | -0.163 |
| hnrnpa0a | 0.231 | gcshb | -0.160 |
| hnrnpa0b | 0.231 | gnpda1 | -0.159 |
| syncrip | 0.227 | COX5B | -0.159 |
| hmgb2b | 0.226 | glud1b | -0.159 |
| marcksb | 0.226 | cdaa | -0.159 |
| hnrnpabb | 0.223 | cubn | -0.158 |
| hdac1 | 0.222 | epdl2 | -0.157 |
| tuba1c | 0.222 | si:dkey-28n18.9 | -0.156 |
| gpm6aa | 0.221 | si:dkey-151g10.6 | -0.156 |
| zc4h2 | 0.220 | aldh6a1 | -0.156 |
| stmn1b | 0.217 | gamt | -0.155 |
| si:ch211-222l21.1 | 0.215 | lrp2a | -0.154 |
| ywhaqb | 0.215 | pdzk1ip1 | -0.153 |
| celf2 | 0.215 | upb1 | -0.153 |
| h2afx1 | 0.213 | si:dkey-103j14.5 | -0.153 |
| tubb5 | 0.213 | sord | -0.153 |
| h3f3d | 0.209 | ldhba | -0.152 |