atonal bHLH transcription factor 1a
ZFIN 
Other cell groups


all cells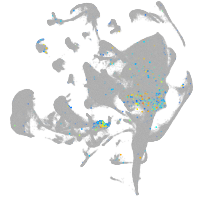 |
blastomeres |
PGCs |
endoderm |
axial mesoderm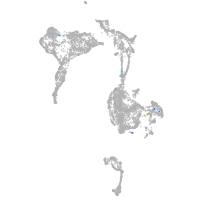 |
muscle 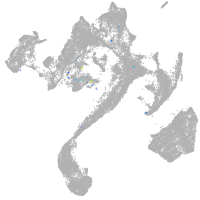 |
mural cells / non-skeletal muscle  |
mesenchyme 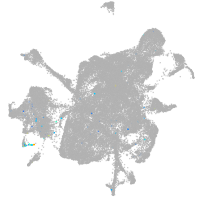 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atoh1b | 0.516 | gpm6ab | -0.086 |
| olig3 | 0.447 | gng3 | -0.086 |
| atoh8 | 0.446 | sncb | -0.085 |
| msx1b | 0.403 | gapdhs | -0.082 |
| wnt10b | 0.324 | rtn1b | -0.080 |
| nog2 | 0.309 | ywhah | -0.078 |
| olig4 | 0.294 | elavl4 | -0.077 |
| fibinb | 0.289 | atp6v0cb | -0.075 |
| msx3 | 0.270 | ckbb | -0.075 |
| cdh6 | 0.246 | ldhba | -0.073 |
| im:7152348 | 0.244 | vamp2 | -0.072 |
| her12 | 0.222 | ywhag2 | -0.072 |
| wnt1 | 0.218 | gpm6aa | -0.071 |
| masp1 | 0.216 | stmn2a | -0.071 |
| lfng | 0.202 | calm2a | -0.070 |
| LOC798783 | 0.200 | stx1b | -0.069 |
| lrrn1 | 0.189 | stxbp1a | -0.069 |
| si:ch73-21g5.7 | 0.186 | zgc:65894 | -0.069 |
| epha2a | 0.186 | atp6v1e1b | -0.068 |
| notch3 | 0.181 | sox4a | -0.067 |
| nr2f6b | 0.179 | snap25a | -0.066 |
| dld | 0.178 | gap43 | -0.066 |
| smad7 | 0.176 | mllt11 | -0.065 |
| cep131 | 0.175 | ywhaz | -0.065 |
| gadd45gb.1 | 0.173 | atpv0e2 | -0.063 |
| cldn5a | 0.172 | si:dkeyp-75h12.5 | -0.063 |
| cdon | 0.171 | tmem59l | -0.063 |
| BX957346.1 | 0.171 | rnasekb | -0.063 |
| nkd1 | 0.169 | atp6v1g1 | -0.063 |
| epha4l | 0.169 | fez1 | -0.063 |
| sertad2a | 0.163 | atp5if1b | -0.063 |
| id1 | 0.161 | tmsb2 | -0.062 |
| XLOC-003689 | 0.160 | id4 | -0.062 |
| fstl1a | 0.160 | tpi1b | -0.062 |
| bmp5 | 0.158 | h2afy2 | -0.062 |




