arrestin domain containing 2
ZFIN 
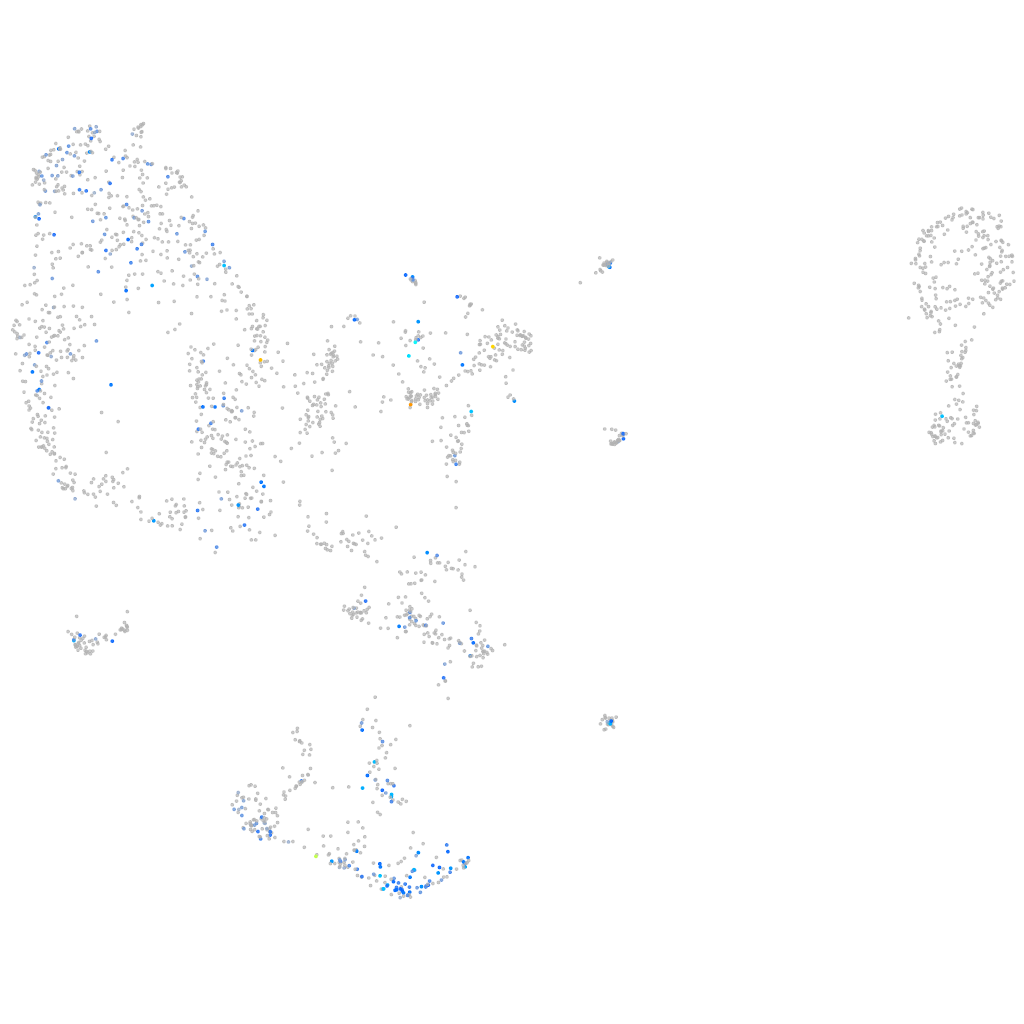
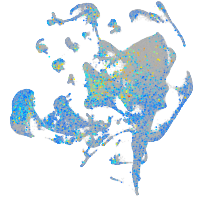
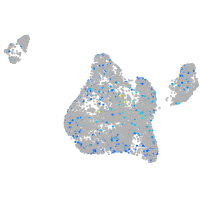
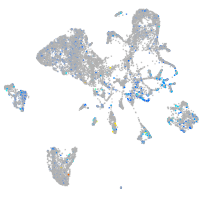
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| vsig8b | 0.279 | ptmab | -0.137 |
| si:dkey-36i7.3 | 0.270 | hnrnpabb | -0.135 |
| rnf183 | 0.267 | hnrnpa1b | -0.127 |
| si:dkey-44g17.6 | 0.266 | wu:fb97g03 | -0.122 |
| si:ch211-284e13.14 | 0.258 | snrpf | -0.121 |
| trim35-12 | 0.254 | seta | -0.119 |
| zgc:158423 | 0.251 | si:ch73-281n10.2 | -0.116 |
| etf1a | 0.249 | syncrip | -0.116 |
| slc9a3.2 | 0.246 | si:ch73-1a9.3 | -0.116 |
| agr2 | 0.246 | eif5a2 | -0.116 |
| s100v2 | 0.243 | nop58 | -0.115 |
| cd63 | 0.243 | nucks1a | -0.115 |
| glud1a | 0.238 | hmga1a | -0.113 |
| soul2 | 0.238 | npm1a | -0.112 |
| atp1a1a.3 | 0.237 | snrpd3l | -0.111 |
| alas1 | 0.237 | snrpe | -0.111 |
| glsb | 0.237 | eif2s2 | -0.111 |
| slc7a8a | 0.228 | snrpg | -0.110 |
| bicdl2l | 0.224 | setb | -0.109 |
| atp1a1a.5 | 0.221 | marcksl1b | -0.108 |
| slc12a10.3 | 0.220 | lig1 | -0.108 |
| mal2 | 0.217 | hspb1 | -0.107 |
| gdpd3a | 0.217 | ptges3b | -0.107 |
| aspg | 0.217 | sumo3b | -0.106 |
| prkag2a | 0.213 | ebna1bp2 | -0.106 |
| hcls1 | 0.212 | fbl | -0.105 |
| BX649516.2 | 0.212 | rbbp4 | -0.104 |
| si:ch211-93f2.1 | 0.210 | snrpd2 | -0.104 |
| nr0b2a | 0.208 | cdx4 | -0.103 |
| tmem176l.4 | 0.207 | cbx3a | -0.103 |
| clcnk | 0.207 | hnrnpa0b | -0.103 |
| zgc:158222 | 0.206 | hnrnpa1a | -0.102 |
| mslnb | 0.205 | pdap1a | -0.102 |
| si:ch211-79k12.1 | 0.205 | si:ch211-137a8.4 | -0.102 |
| tmem72 | 0.204 | hnrnpa0a | -0.101 |