Rho guanine nucleotide exchange factor (GEF) 1a
ZFIN 
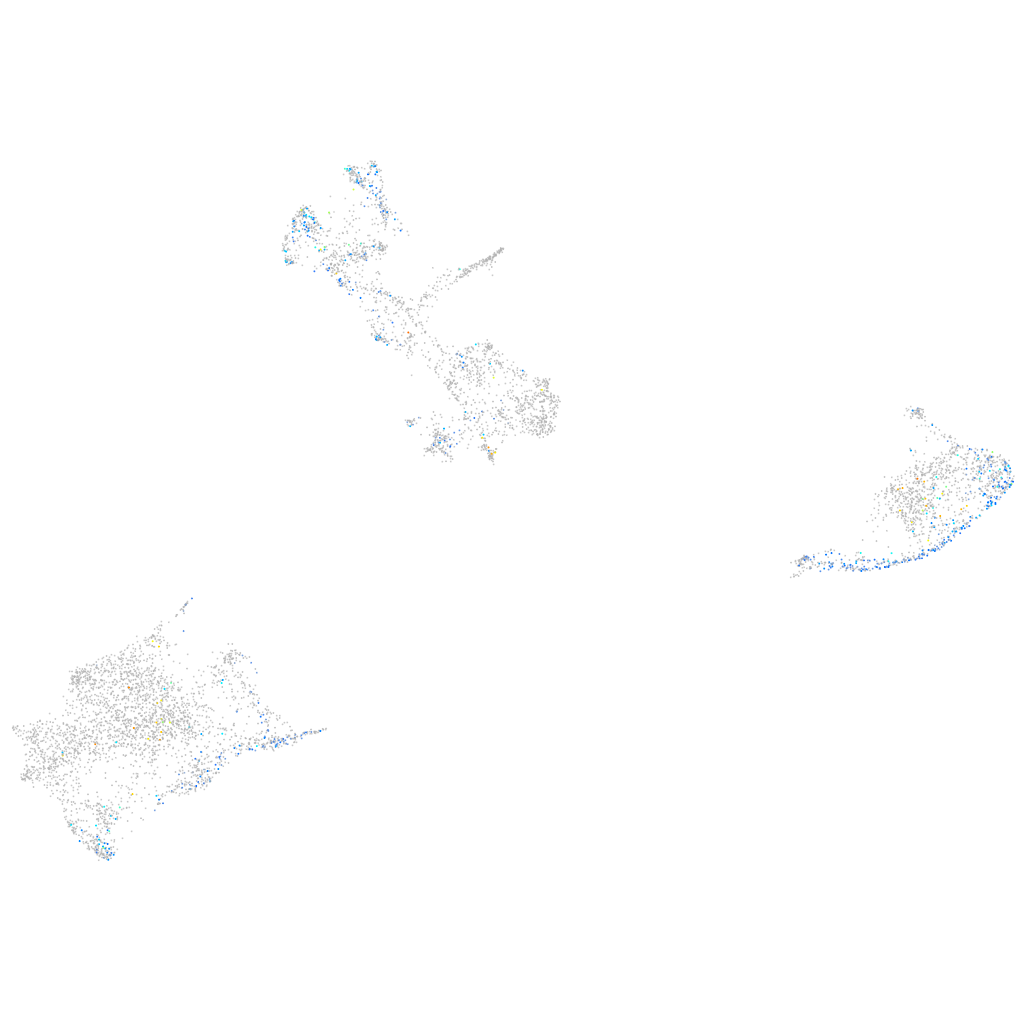
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc45a2 | 0.158 | CABZ01021592.1 | -0.124 |
| atp11a | 0.150 | si:dkey-251i10.2 | -0.109 |
| SPAG9 | 0.144 | uraha | -0.086 |
| slc37a2 | 0.142 | si:ch211-222l21.1 | -0.075 |
| kita | 0.138 | tmem130 | -0.071 |
| slc29a3 | 0.135 | hbbe1.3 | -0.071 |
| slc3a2a | 0.133 | tuba1c | -0.070 |
| tspan36 | 0.132 | hbae3 | -0.070 |
| smim29 | 0.132 | krt4 | -0.067 |
| oca2 | 0.131 | elavl3 | -0.067 |
| atp6v0ca | 0.130 | ptmaa | -0.066 |
| dct | 0.130 | hbae1.1 | -0.066 |
| tyrp1a | 0.129 | si:ch211-251b21.1 | -0.064 |
| tyr | 0.128 | tmsb | -0.063 |
| pmela | 0.128 | gch2 | -0.062 |
| arhgef33 | 0.126 | pvalb2 | -0.061 |
| msx1b | 0.125 | hmgb1b | -0.060 |
| rab38 | 0.124 | aqp3a | -0.059 |
| slc24a5 | 0.124 | oacyl | -0.058 |
| zgc:91968 | 0.123 | hbbe1.1 | -0.058 |
| si:zfos-943e10.1 | 0.123 | bco1 | -0.058 |
| gadd45ba | 0.123 | gng3 | -0.058 |
| sptlc2a | 0.123 | aox5 | -0.058 |
| hipk3b | 0.122 | hmgb3a | -0.057 |
| mtbl | 0.122 | sult1st1 | -0.057 |
| slc16a1b | 0.122 | stmn1b | -0.056 |
| map7a | 0.122 | marcksl1a | -0.056 |
| lamp1a | 0.121 | actc1b | -0.056 |
| rab32a | 0.121 | slc22a7a | -0.056 |
| tyrp1b | 0.121 | ckma | -0.056 |
| trpm1b | 0.120 | pvalb1 | -0.055 |
| AL935199.1 | 0.120 | hmgn2 | -0.055 |
| mitfa | 0.119 | si:dkey-73n8.3 | -0.054 |
| id2a | 0.119 | prdx5 | -0.053 |
| pla2g15 | 0.119 | paics | -0.053 |