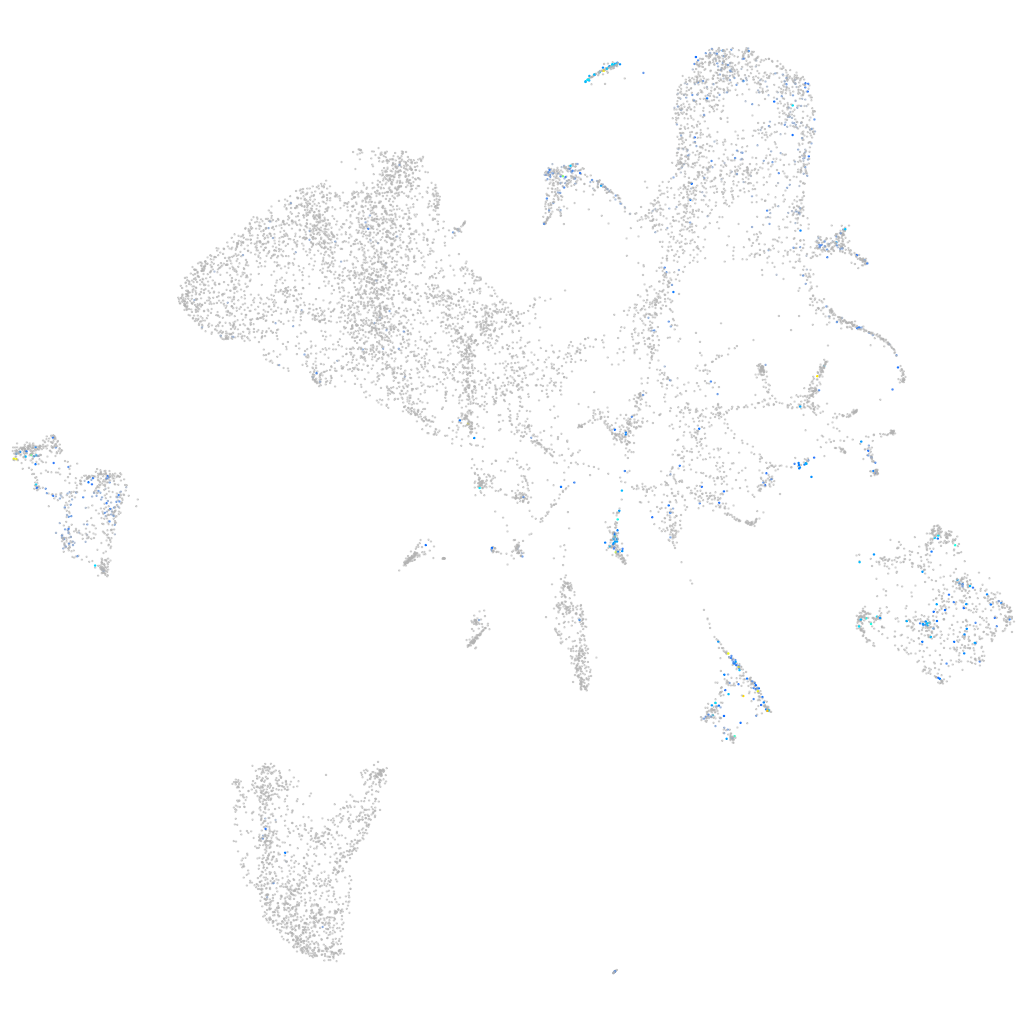
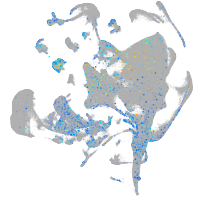
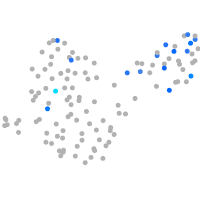
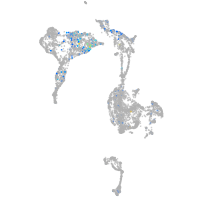
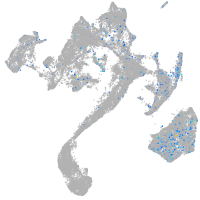
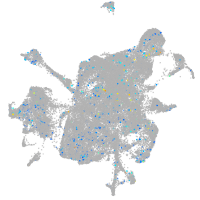
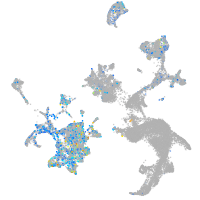
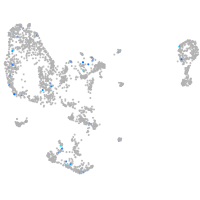
Rho guanine nucleotide exchange factor (GEF) 1a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-234i14.2 | 0.179 | bhmt | -0.157 |
| adcyap1a | 0.178 | ahcy | -0.155 |
| hbegfb | 0.176 | gamt | -0.147 |
| lmx1ba | 0.168 | aqp12 | -0.147 |
| degs2 | 0.166 | gatm | -0.137 |
| penka | 0.157 | fabp3 | -0.137 |
| elovl1b | 0.151 | mat1a | -0.137 |
| myofl | 0.151 | agxtb | -0.130 |
| ier2b | 0.150 | apoa1b | -0.129 |
| fthl27 | 0.150 | gapdh | -0.129 |
| slc7a14a | 0.149 | apoa2 | -0.125 |
| si:dkey-208c12.2 | 0.148 | agxta | -0.122 |
| ahnak | 0.148 | kng1 | -0.121 |
| gna15.1 | 0.148 | serpina1 | -0.119 |
| trpa1b | 0.148 | zgc:123103 | -0.118 |
| zgc:92380 | 0.148 | serpinf2b | -0.118 |
| abca12 | 0.147 | serpina1l | -0.117 |
| zgc:101731 | 0.146 | vtnb | -0.117 |
| gapdhs | 0.144 | ttc36 | -0.116 |
| CABZ01055365.1 | 0.143 | grhprb | -0.116 |
| anxa3b | 0.142 | ftcd | -0.116 |
| znf185 | 0.141 | apom | -0.115 |
| zgc:63831 | 0.138 | fgg | -0.115 |
| sim1b | 0.138 | hao1 | -0.115 |
| f3a | 0.138 | rbp2b | -0.115 |
| cabp2b | 0.138 | fabp10a | -0.115 |
| sgms1 | 0.137 | hpda | -0.114 |
| wu:fj05g07 | 0.137 | cfhl4 | -0.114 |
| cygb1 | 0.136 | ces2 | -0.113 |
| stm | 0.136 | ambp | -0.113 |
| actb1 | 0.135 | fgb | -0.113 |
| epcam | 0.135 | fetub | -0.113 |
| actn1 | 0.135 | hspe1 | -0.112 |
| tspan15 | 0.135 | shmt1 | -0.111 |
| loxl5a | 0.134 | LOC110437731 | -0.111 |