Rho GTPase activating protein 42a
ZFIN 
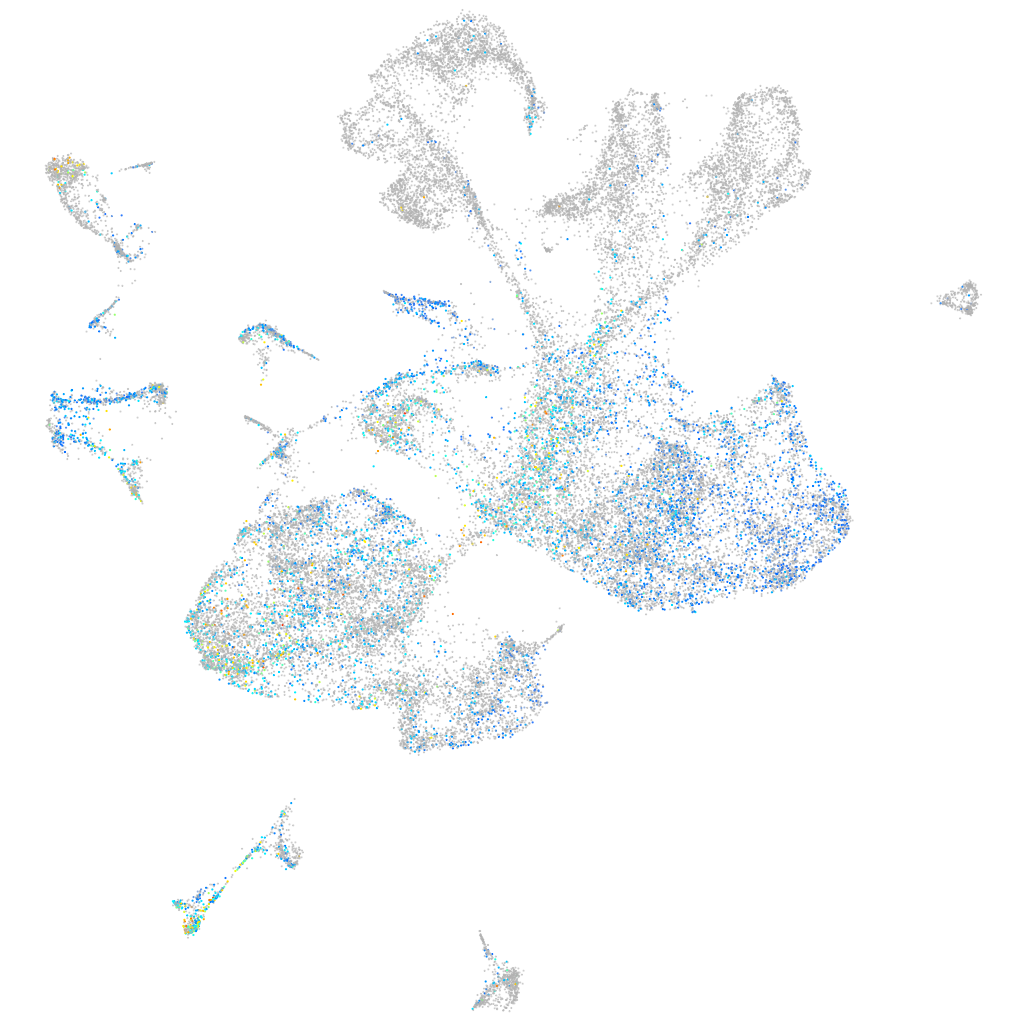
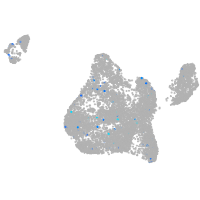
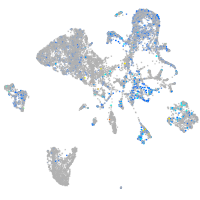
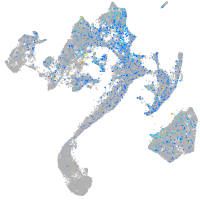
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-335m24.2 | 0.171 | snap25a | -0.136 |
| qki2 | 0.163 | stxbp1a | -0.136 |
| cd63 | 0.163 | sncb | -0.135 |
| cd82a | 0.156 | stx1b | -0.134 |
| cd226 | 0.155 | elavl4 | -0.134 |
| vamp3 | 0.142 | gng3 | -0.130 |
| lmo7a | 0.137 | cplx2 | -0.127 |
| si:dkey-204f11.64 | 0.136 | syt2a | -0.124 |
| plp1b | 0.132 | rtn1b | -0.123 |
| si:ch211-286b5.5 | 0.130 | nsg2 | -0.123 |
| sept8b | 0.130 | tmem59l | -0.118 |
| npc2 | 0.126 | stmn2a | -0.118 |
| si:dkey-200l5.4 | 0.123 | gng2 | -0.117 |
| id1 | 0.122 | ywhag2 | -0.116 |
| crp1 | 0.121 | cplx2l | -0.116 |
| swap70b | 0.121 | rbfox1 | -0.115 |
| c7b | 0.120 | tmsb | -0.114 |
| sdcbp2 | 0.120 | onecut1 | -0.114 |
| BX000438.1 | 0.120 | ywhah | -0.114 |
| wls | 0.119 | vamp2 | -0.113 |
| hepacama | 0.118 | ptmaa | -0.113 |
| glipr2l | 0.118 | gap43 | -0.112 |
| pgrmc1 | 0.118 | sncgb | -0.111 |
| slc31a2 | 0.117 | csdc2a | -0.110 |
| sox2 | 0.117 | elavl3 | -0.109 |
| cldn19 | 0.117 | myt1b | -0.109 |
| crip3 | 0.116 | atp6v0cb | -0.108 |
| ptprfb | 0.115 | id4 | -0.107 |
| atp1b4 | 0.115 | si:dkey-276j7.1 | -0.106 |
| sparc | 0.113 | atp1a3a | -0.106 |
| XLOC-003690 | 0.113 | rtn1a | -0.105 |
| CABZ01059119.1 | 0.112 | eno2 | -0.105 |
| csrp1b | 0.111 | ndrg4 | -0.103 |
| mag | 0.111 | rbfox3a | -0.102 |
| si:ch211-165b10.3 | 0.110 | jagn1a | -0.101 |