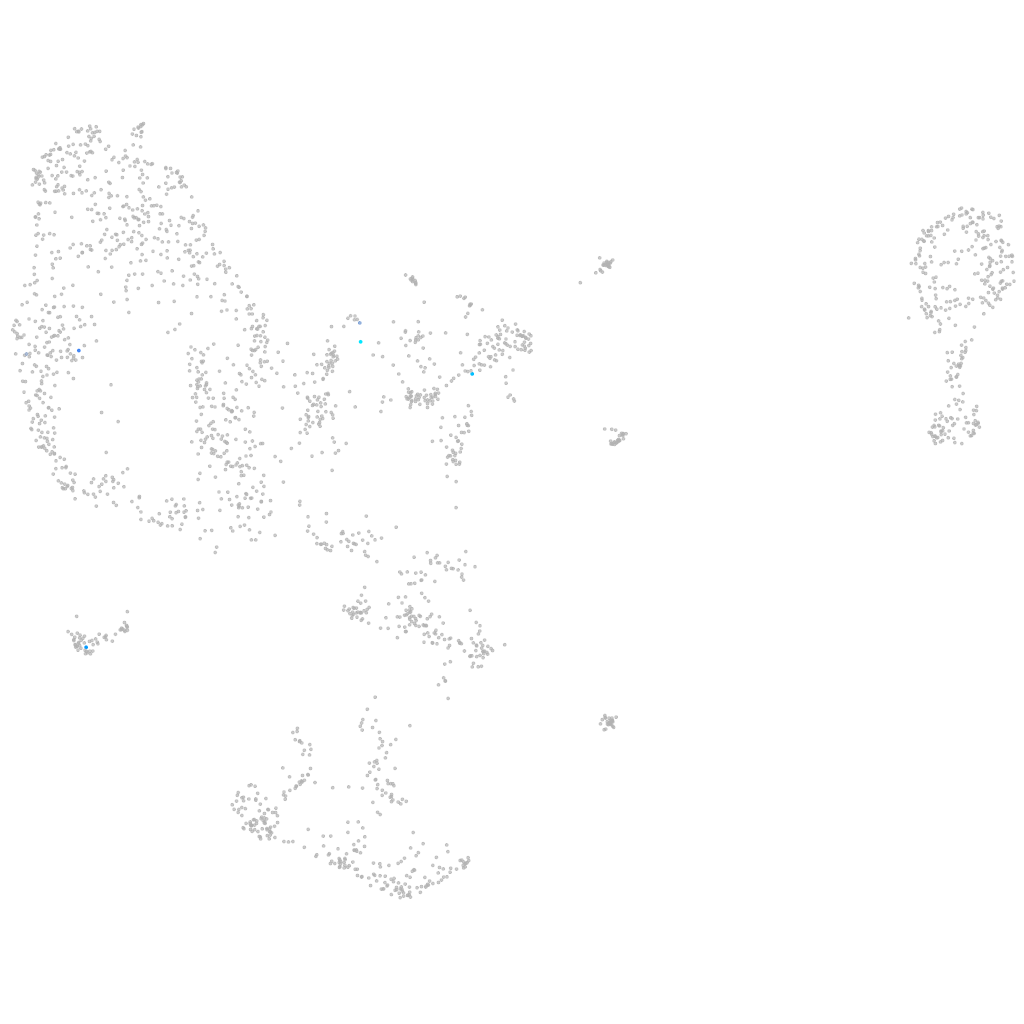
Rho GTPase activating protein 20b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 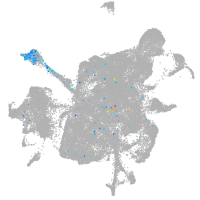 |
hematopoietic / vasculature 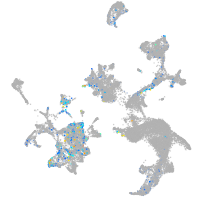 |
pronephros  |
neural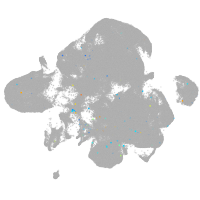 |
spinal cord / glia |
eye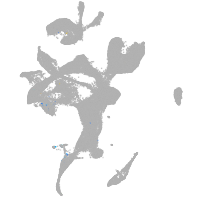 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| MINAR1 | 0.545 | ndufa8 | -0.058 |
| LOC100332458 | 0.506 | eif3ba | -0.056 |
| mrc1a | 0.449 | ccng1 | -0.053 |
| RF00566 | 0.439 | skp1 | -0.051 |
| gpib | 0.404 | arpc3 | -0.048 |
| XLOC-043113 | 0.389 | cct4 | -0.048 |
| CR759810.1 | 0.367 | pfdn4 | -0.047 |
| syne2a | 0.357 | ndrg3a | -0.047 |
| LOC103909511 | 0.347 | cct6a | -0.047 |
| aqp7 | 0.335 | cox8a | -0.045 |
| si:dkeyp-97a10.2 | 0.331 | psma4 | -0.045 |
| flt4 | 0.325 | mrpl54 | -0.045 |
| myct1a | 0.316 | timm8b | -0.043 |
| zgc:195282 | 0.316 | GCA | -0.043 |
| ablim1a | 0.314 | eif5 | -0.043 |
| ecscr | 0.306 | tmed2 | -0.043 |
| si:ch211-145b13.6 | 0.299 | ndufs2 | -0.043 |
| tie1 | 0.291 | vamp3 | -0.043 |
| CU929451.2 | 0.287 | pa2g4b | -0.043 |
| frmpd1b | 0.285 | hadhb | -0.042 |
| pdlim5a | 0.282 | glrx5 | -0.042 |
| CU459093.1 | 0.278 | ola1 | -0.042 |
| si:ch211-66e2.3 | 0.277 | sdcbp2 | -0.042 |
| tll1 | 0.271 | atp6v0ca | -0.042 |
| FP102309.1 | 0.268 | ddt | -0.042 |
| scube3 | 0.266 | ehd1b | -0.042 |
| si:dkey-5n18.1 | 0.259 | ube2ib | -0.041 |
| BX248521.2 | 0.257 | ndufa7 | -0.040 |
| slco2b1 | 0.254 | timm9 | -0.040 |
| LOC101882049 | 0.253 | zgc:91910 | -0.040 |
| si:ch211-161c3.6 | 0.249 | si:ch211-286b5.5 | -0.040 |
| ttc29 | 0.247 | dldh | -0.040 |
| si:dkey-92i15.4 | 0.246 | eef1b2 | -0.039 |
| XLOC-011909 | 0.246 | phb2b | -0.039 |
| kcnk10b | 0.243 | rpl6 | -0.039 |