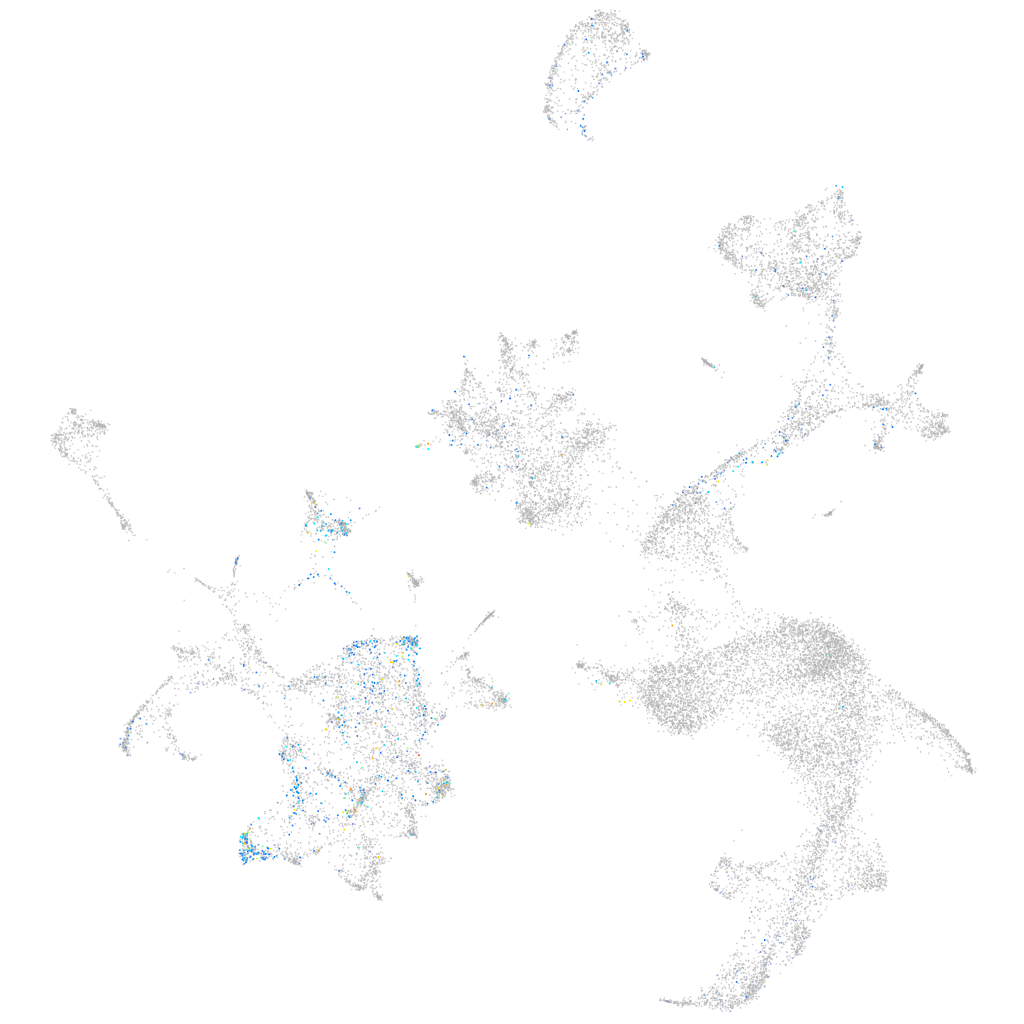
Rho GTPase activating protein 20b
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory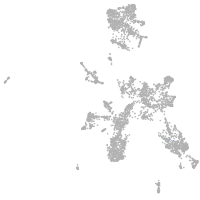 |
otic / lateral line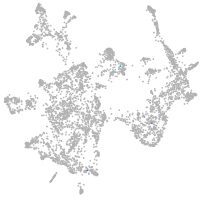 |
epidermis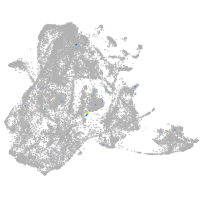 |
periderm |
fin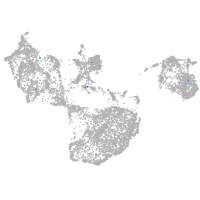 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cdh5 | 0.205 | hbbe2 | -0.135 |
| itga1 | 0.192 | hbae1.1 | -0.132 |
| krt8 | 0.192 | hbae3 | -0.131 |
| pecam1 | 0.189 | hbae1.3 | -0.129 |
| col4a1 | 0.188 | hbbe1.3 | -0.129 |
| clec14a | 0.188 | hbbe1.1 | -0.126 |
| si:ch211-156j16.1 | 0.183 | cahz | -0.123 |
| krt18a.1 | 0.180 | prdx2 | -0.119 |
| cldn5b | 0.179 | fth1a | -0.118 |
| tie1 | 0.178 | hemgn | -0.117 |
| cav1 | 0.177 | hbbe1.2 | -0.112 |
| si:ch211-145b13.6 | 0.177 | si:ch211-250g4.3 | -0.107 |
| tspan4b | 0.177 | alas2 | -0.105 |
| eppk1 | 0.172 | gpx1a | -0.102 |
| cavin2b | 0.171 | nt5c2l1 | -0.101 |
| tpm4a | 0.169 | blvrb | -0.101 |
| myl9a | 0.167 | creg1 | -0.097 |
| mcamb | 0.167 | slc4a1a | -0.096 |
| flna | 0.167 | si:ch211-207c6.2 | -0.096 |
| cd81a | 0.167 | zgc:163057 | -0.092 |
| plvapb | 0.167 | tspo | -0.091 |
| gpr182 | 0.165 | epb41b | -0.089 |
| ahnak | 0.164 | tmod4 | -0.088 |
| fgd5a | 0.162 | nmt1b | -0.084 |
| kdrl | 0.161 | vdac2 | -0.080 |
| igfbp7 | 0.161 | zgc:56095 | -0.079 |
| ecscr | 0.159 | hbae5 | -0.074 |
| kdr | 0.157 | si:ch211-227m13.1 | -0.073 |
| ehd2b | 0.157 | plac8l1 | -0.072 |
| calcrla | 0.156 | add2 | -0.070 |
| she | 0.156 | si:dkey-240n22.3 | -0.069 |
| limch1b | 0.156 | rfesd | -0.063 |
| fli1a | 0.154 | selenoj | -0.063 |
| si:dkeyp-97a10.2 | 0.154 | klf1 | -0.062 |
| cpn1 | 0.152 | TCIM (1 of many) | -0.062 |