apolipoprotein A-IV a
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 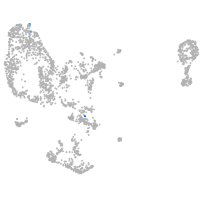 |
neural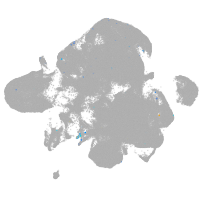 |
spinal cord / glia |
eye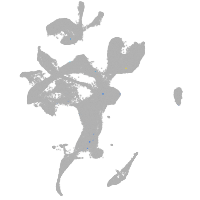 |
taste / olfactory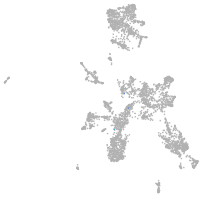 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01081535.1 | 0.131 | ptmab | -0.022 |
| si:ch211-113d11.6 | 0.128 | tubb2b | -0.015 |
| c1r | 0.127 | marcksl1b | -0.014 |
| apoa4b.2 | 0.127 | hmgb1b | -0.014 |
| AL831745.1 | 0.122 | ywhabl | -0.012 |
| LOC103908636 | 0.119 | hnrnpa0a | -0.012 |
| zanl | 0.117 | hmgb1a | -0.011 |
| wu:fa56d06 | 0.115 | hmgn7 | -0.011 |
| zgc:153968 | 0.113 | tmeff1b | -0.011 |
| si:ch211-133l5.7 | 0.111 | tuba1c | -0.011 |
| LOC110437835 | 0.110 | nova2 | -0.011 |
| ace2 | 0.110 | hnrnpaba | -0.011 |
| enpp7.1 | 0.109 | hmgn6 | -0.011 |
| abcc2 | 0.108 | si:ch73-1a9.3 | -0.010 |
| myo7bb | 0.107 | tox | -0.010 |
| chia.2 | 0.106 | hmgb3a | -0.010 |
| cyp3a65 | 0.105 | rtn1a | -0.010 |
| ccdc17 | 0.100 | calm1a | -0.010 |
| entpd8 | 0.099 | snap25a | -0.010 |
| gpr39 | 0.099 | si:dkey-276j7.1 | -0.010 |
| plac8.1 | 0.098 | cadm3 | -0.010 |
| neu3.3 | 0.098 | ywhag2 | -0.010 |
| dgat2 | 0.098 | stmn1b | -0.010 |
| zgc:172079 | 0.098 | sncb | -0.010 |
| XLOC-012620 | 0.095 | elavl4 | -0.010 |
| slc6a19a.2 | 0.094 | cnrip1a | -0.010 |
| si:ch211-196f2.3 | 0.094 | ptmaa | -0.010 |
| sult1st2 | 0.093 | khdrbs1a | -0.009 |
| zgc:158846 | 0.093 | cpeb4b | -0.009 |
| CABZ01088484.1 | 0.091 | dnajc5aa | -0.009 |
| apoea | 0.089 | elavl3 | -0.009 |
| zgc:112146 | 0.089 | hnrnpa0b | -0.009 |
| vil1 | 0.089 | gng3 | -0.009 |
| chs1 | 0.088 | stx1b | -0.009 |
| slc15a1b | 0.087 | sypb | -0.009 |




