apelin
ZFIN 
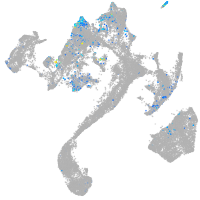
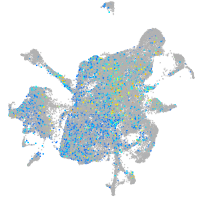
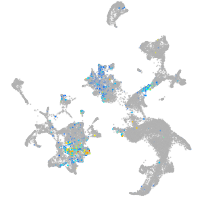
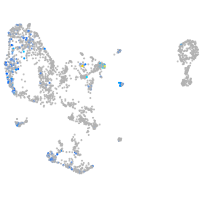
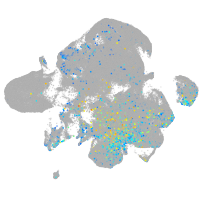
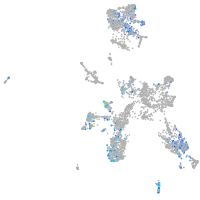
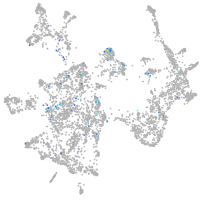
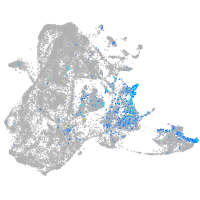
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch73-233f7.4 | 0.302 | actb2 | -0.068 |
| BX276128.1 | 0.291 | gpr143 | -0.058 |
| eif3bb | 0.265 | rpl12 | -0.058 |
| CABZ01117780.1 | 0.244 | syngr1a | -0.057 |
| MINAR1 | 0.232 | smim29 | -0.056 |
| XLOC-017471 | 0.232 | tspan36 | -0.054 |
| dzank1 | 0.222 | pfn1 | -0.053 |
| XLOC-029547 | 0.218 | si:dkey-151g10.6 | -0.051 |
| BX927333.1 | 0.217 | rab38 | -0.051 |
| LOC101883723 | 0.216 | trpm1b | -0.050 |
| sim2.1 | 0.215 | C12orf75 | -0.049 |
| sstr1b | 0.211 | rabl6b | -0.047 |
| slitrk3b | 0.205 | slc22a7a | -0.047 |
| lrrc38b | 0.202 | rps19 | -0.046 |
| XLOC-011126 | 0.200 | rps23 | -0.046 |
| caprin2 | 0.199 | si:dkey-21a6.5 | -0.045 |
| grxcr1a.1 | 0.199 | glulb | -0.045 |
| cacna1ab | 0.198 | eno3 | -0.044 |
| slc17a6a | 0.198 | pfas | -0.044 |
| ccdc120 | 0.198 | akr1b1 | -0.043 |
| nptx2a | 0.198 | slc2a15a | -0.043 |
| bsnb | 0.196 | myo5aa | -0.043 |
| guca1e | 0.190 | gart | -0.043 |
| kcns3a | 0.186 | bace2 | -0.043 |
| si:dkey-4e4.1 | 0.185 | pttg1ipb | -0.043 |
| kcna1a | 0.184 | glrx | -0.042 |
| zgc:193790 | 0.184 | uraha | -0.042 |
| slc6a1a | 0.182 | rpl11 | -0.042 |
| scg2b | 0.180 | tfap2e | -0.042 |
| bmp16 | 0.174 | CABZ01032488.1 | -0.042 |
| foxf2b | 0.172 | opn5 | -0.042 |
| LOC101887100 | 0.171 | impdh1b | -0.041 |
| plpp2a | 0.170 | rplp2l | -0.041 |
| CABZ01039096.1 | 0.170 | ppat | -0.041 |
| si:ch1073-186i23.1 | 0.168 | cd164 | -0.041 |