annexin A6
ZFIN 
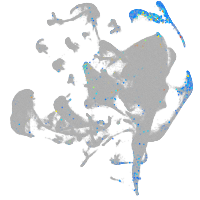
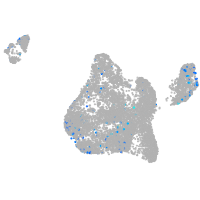
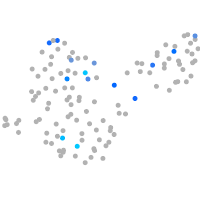
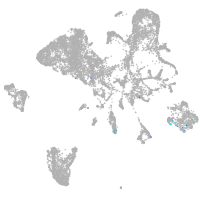
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cavin4a | 0.401 | marcksl1a | -0.080 |
| cav3 | 0.400 | hmgb1b | -0.078 |
| srl | 0.392 | gpm6aa | -0.073 |
| CABZ01078594.1 | 0.390 | nova2 | -0.071 |
| casq2 | 0.387 | tuba1c | -0.071 |
| cavin4b | 0.386 | ywhaqb | -0.070 |
| zgc:158296 | 0.380 | si:ch211-288g17.3 | -0.069 |
| actn3b | 0.379 | mdkb | -0.065 |
| tmem182a | 0.379 | gpm6ab | -0.064 |
| ldb3b | 0.378 | chd4a | -0.063 |
| trdn | 0.377 | rnasekb | -0.063 |
| pgam2 | 0.376 | ckbb | -0.060 |
| aldoab | 0.375 | gapdhs | -0.060 |
| hhatla | 0.373 | gnb1b | -0.060 |
| desma | 0.372 | h2afva | -0.060 |
| neb | 0.368 | atp6v0cb | -0.059 |
| stac3 | 0.367 | elavl3 | -0.059 |
| tmem38a | 0.367 | hmgb3a | -0.059 |
| myom1a | 0.366 | stmn1b | -0.059 |
| klhl43 | 0.366 | tmeff1b | -0.059 |
| dhrs7cb | 0.363 | zc4h2 | -0.059 |
| prr33 | 0.362 | si:ch211-137a8.4 | -0.058 |
| si:ch211-266g18.10 | 0.362 | si:dkey-276j7.1 | -0.057 |
| CABZ01072309.1 | 0.360 | hmgn3 | -0.056 |
| CU681836.1 | 0.359 | tuba1a | -0.056 |
| cacna1sb | 0.358 | marcksb | -0.055 |
| casq1b | 0.357 | tubb5 | -0.055 |
| actn3a | 0.356 | cspg5a | -0.053 |
| mybphb | 0.355 | fam168a | -0.053 |
| si:dkey-211f22.5 | 0.355 | hdac1 | -0.053 |
| CR376766.2 | 0.354 | atrx | -0.052 |
| klhl31 | 0.354 | epb41a | -0.052 |
| ldb3a | 0.354 | cadm3 | -0.051 |
| tnnt3a | 0.352 | calm3b | -0.051 |
| smyd1a | 0.350 | seta | -0.051 |