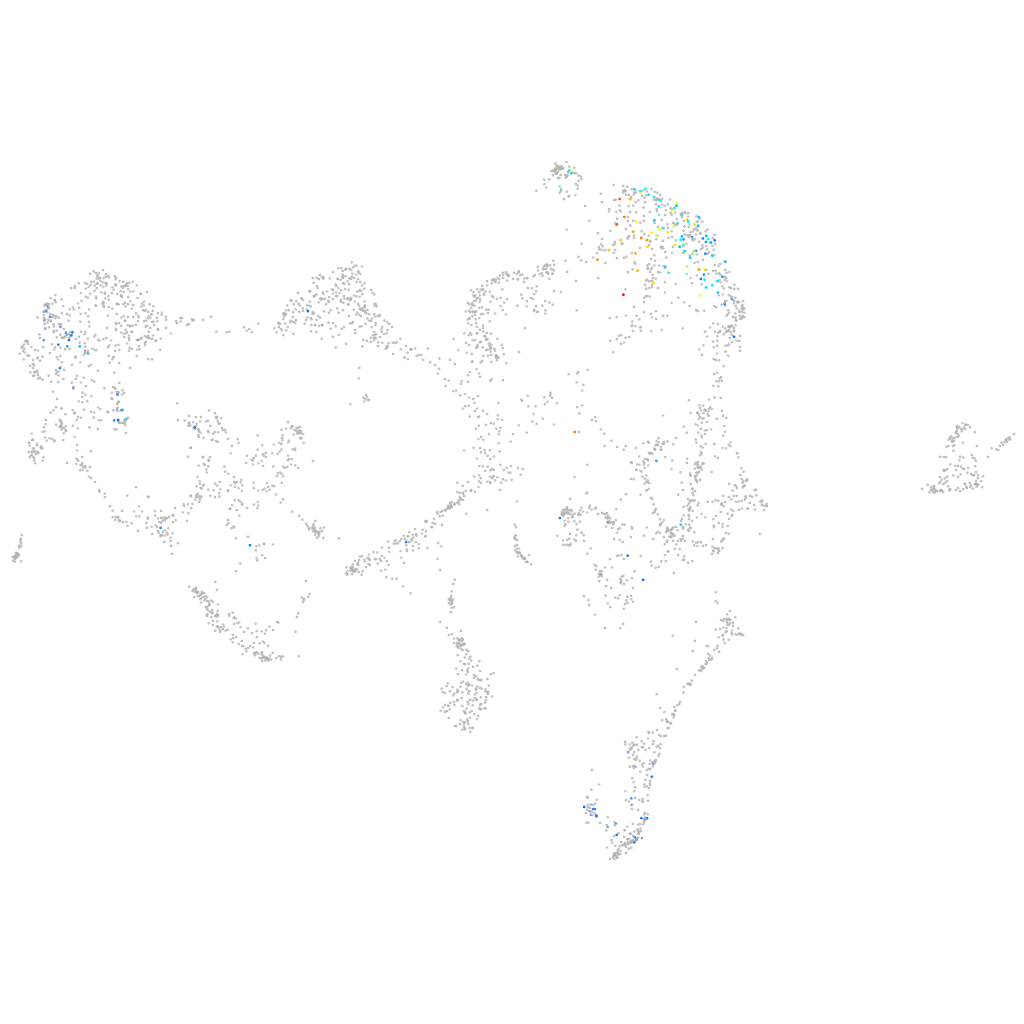
ankyrin repeat and death domain containing 1B
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 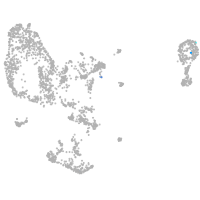 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis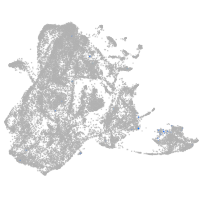 |
periderm |
fin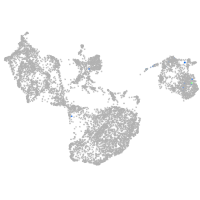 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC110438378 | 0.370 | atp5mc3b | -0.223 |
| muc5.2 | 0.364 | si:dkey-33i11.4 | -0.219 |
| si:dkey-65b12.6 | 0.334 | COX5B | -0.203 |
| muc5.1 | 0.324 | cox7a2a | -0.202 |
| atp2a3 | 0.318 | atp5mc1 | -0.198 |
| npdc1a | 0.298 | atp5f1b | -0.191 |
| stard10 | 0.290 | atp5if1a | -0.188 |
| b3gnt7 | 0.277 | atp1b1b | -0.186 |
| gng13b | 0.267 | cox7b | -0.183 |
| zgc:112994 | 0.264 | cox6a1 | -0.180 |
| sytl2b | 0.263 | atp5mf | -0.177 |
| calm1b | 0.255 | idh2 | -0.177 |
| si:dkey-1h24.2 | 0.255 | uqcrh | -0.175 |
| p2rx1 | 0.252 | atp5f1e | -0.173 |
| rap1gap | 0.244 | slc25a5 | -0.172 |
| zgc:92380 | 0.242 | cox6b2 | -0.171 |
| gpx3 | 0.241 | atp5pd | -0.171 |
| sytl4 | 0.239 | atp5l | -0.170 |
| agr2 | 0.239 | prdx2 | -0.169 |
| itpr3 | 0.236 | atp5fa1 | -0.169 |
| cmah | 0.233 | uqcrc1 | -0.168 |
| kita | 0.230 | uqcrq | -0.166 |
| si:ch73-306e8.2 | 0.225 | ndufs5 | -0.165 |
| fer1l6 | 0.220 | atp5f1c | -0.165 |
| rgs2 | 0.219 | ndrg3a | -0.164 |
| slc46a3 | 0.217 | zgc:193541 | -0.164 |
| sytl1 | 0.215 | uqcrfs1 | -0.163 |
| crip1 | 0.215 | ndufab1a | -0.163 |
| ern2 | 0.213 | nqo1 | -0.162 |
| spaca4l | 0.213 | atp5f1d | -0.161 |
| lad1 | 0.212 | atp5meb | -0.161 |
| ponzr6 | 0.211 | atp5md | -0.160 |
| aldob | 0.209 | cox4i1 | -0.160 |
| ucp2 | 0.209 | mpc1 | -0.159 |
| cd63 | 0.208 | mdh2 | -0.158 |