angiopoietin 2a
ZFIN 
Other cell groups


all cells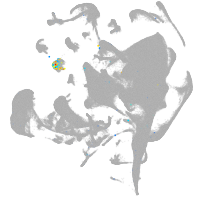 |
blastomeres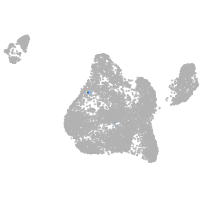 |
PGCs |
endoderm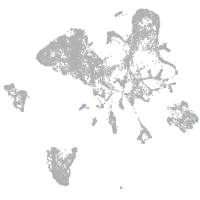 |
axial mesoderm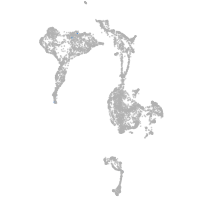 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 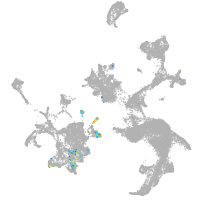 |
pronephros 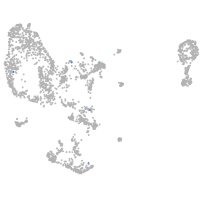 |
neural |
spinal cord / glia |
eye |
taste / olfactory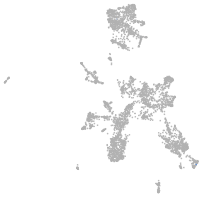 |
otic / lateral line |
epidermis |
periderm |
fin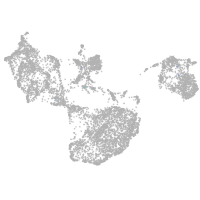 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lyve1a | 0.877 | xbp1 | -0.023 |
| tnfaip2b | 0.660 | pfn1 | -0.019 |
| notchl | 0.632 | egfl6 | -0.019 |
| kdr | 0.545 | tpd52l2b | -0.019 |
| acyp2 | 0.537 | ndufa6 | -0.019 |
| gypc | 0.485 | ywhabb | -0.019 |
| CT033796.1 | 0.463 | msx2b | -0.018 |
| stab1 | 0.463 | serp1 | -0.018 |
| myct1a | 0.449 | tmsb1 | -0.018 |
| rasip1 | 0.446 | socs3a | -0.018 |
| mrc1a | 0.396 | krtcap2 | -0.018 |
| LOC101885919 | 0.395 | dnase1l4.1 | -0.018 |
| plvapb | 0.393 | XLOC-035413 | -0.018 |
| mfng | 0.387 | hdlbpa | -0.017 |
| si:ch211-33e4.2 | 0.359 | bcam | -0.017 |
| ecscr | 0.343 | midn | -0.017 |
| gpr182 | 0.324 | ssr2 | -0.017 |
| LOC101882178 | 0.321 | cfl1l | -0.017 |
| CABZ01029422.1 | 0.316 | oaz1b | -0.017 |
| CU927890.1 | 0.301 | bsg | -0.017 |
| fgd5a | 0.289 | pak2a | -0.017 |
| pecam1 | 0.272 | cldni | -0.016 |
| clec14a | 0.272 | wdr1 | -0.016 |
| adgre5b.2 | 0.271 | jdp2b | -0.016 |
| itln2 | 0.265 | thy1 | -0.016 |
| BX005085.1 | 0.259 | apoeb | -0.016 |
| serpind1 | 0.256 | mapk12a | -0.016 |
| yrk | 0.250 | tax1bp3 | -0.016 |
| dysf | 0.246 | epcam | -0.016 |
| flt4 | 0.244 | cldn1 | -0.016 |
| klhl4 | 0.242 | fosb | -0.016 |
| znf1163 | 0.239 | cotl1 | -0.016 |
| si:ch73-334d15.2 | 0.223 | bzw1b | -0.015 |
| BX324212.2 | 0.219 | tp63 | -0.015 |
| si:ch211-105f12.2 | 0.219 | s100a10b | -0.015 |




