aryl hydrocarbon receptor interacting protein-like 1
ZFIN 
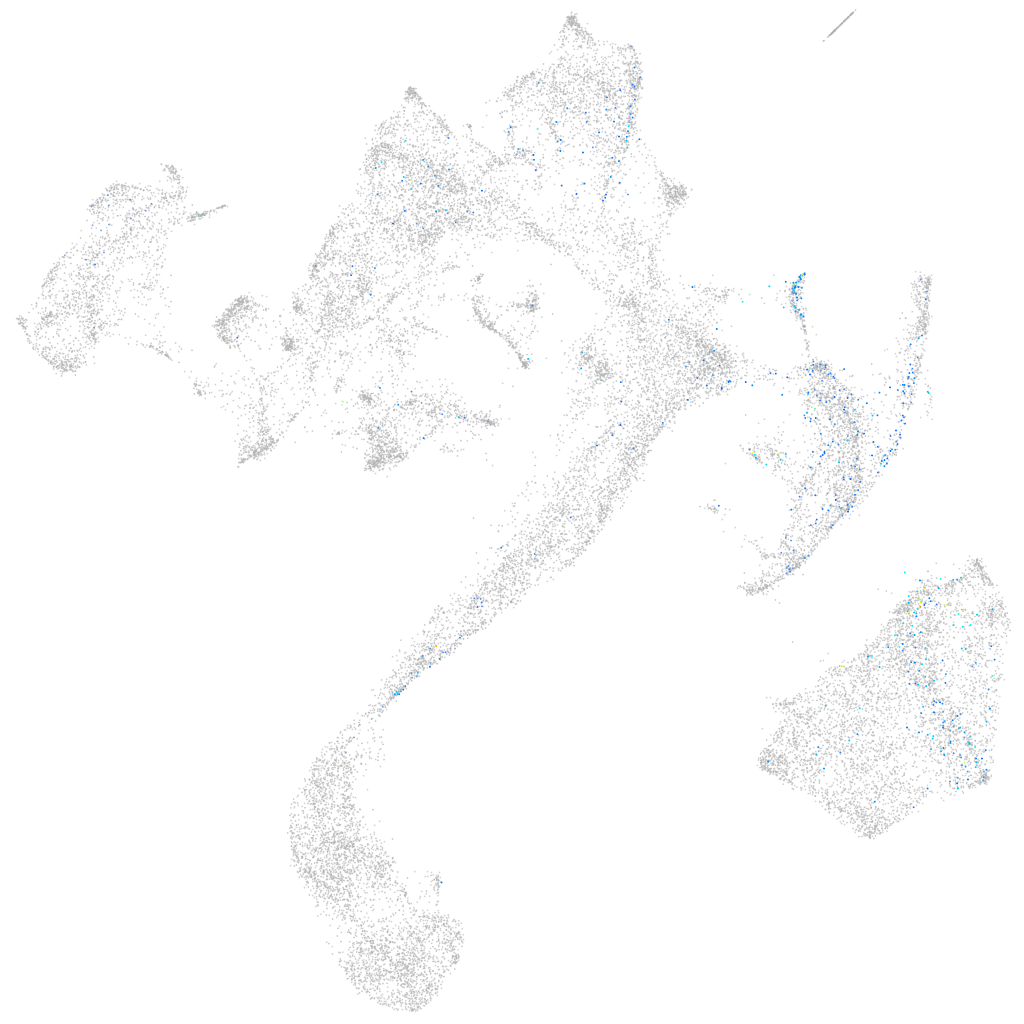
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| msgn1 | 0.144 | actc1b | -0.064 |
| apoc1 | 0.124 | fabp3 | -0.063 |
| itm2cb | 0.119 | ttn.2 | -0.059 |
| tbx16l | 0.117 | pabpc4 | -0.058 |
| hes6 | 0.117 | ak1 | -0.057 |
| tbx16 | 0.114 | atp2a1 | -0.056 |
| aplnra | 0.109 | ckma | -0.056 |
| nid2a | 0.109 | pvalb1 | -0.055 |
| her7 | 0.106 | celf2 | -0.055 |
| alpi.1 | 0.105 | ckmb | -0.055 |
| apoeb | 0.104 | tmem38a | -0.055 |
| prl2 | 0.103 | gamt | -0.055 |
| pcdh8 | 0.102 | nme2b.2 | -0.053 |
| efnb2b | 0.100 | pvalb2 | -0.053 |
| hoxa13a | 0.098 | neb | -0.053 |
| her1 | 0.095 | zgc:158463 | -0.052 |
| cx43.4 | 0.095 | mylpfa | -0.052 |
| inka1b | 0.094 | eno1a | -0.052 |
| cxcl12b | 0.091 | aldoab | -0.051 |
| snai1a | 0.091 | tnnc2 | -0.050 |
| im:7138239 | 0.089 | bhmt | -0.050 |
| rbm38 | 0.088 | si:dkey-16p21.8 | -0.050 |
| vox | 0.086 | gapdh | -0.050 |
| asph | 0.085 | myl1 | -0.049 |
| ved | 0.085 | si:ch211-266g18.10 | -0.049 |
| si:ch73-281n10.2 | 0.085 | sparc | -0.049 |
| cdx4 | 0.083 | tnnt3b | -0.049 |
| zgc:153760 | 0.083 | ldb3a | -0.049 |
| tbx6 | 0.083 | eef1da | -0.049 |
| tuba8l2 | 0.083 | actn3a | -0.049 |
| mespaa | 0.083 | rpl37 | -0.049 |
| cmtm6 | 0.083 | ldb3b | -0.048 |
| si:cabz01036006.1 | 0.083 | eif4a1b | -0.048 |
| ddit4 | 0.082 | si:ch73-367p23.2 | -0.048 |
| hoxb13a | 0.081 | tmod4 | -0.048 |