angiogenic factor with G patch and FHA domains 1
ZFIN 
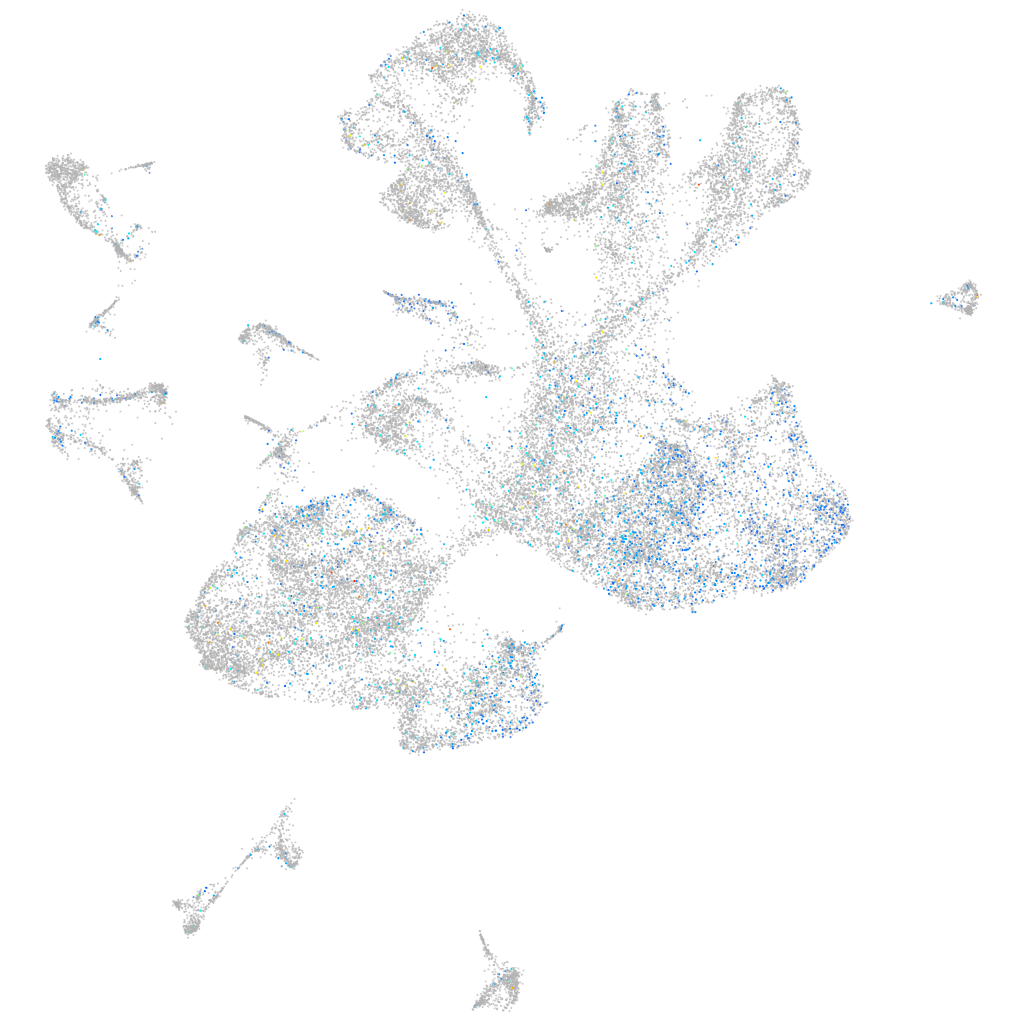
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hmgb2a | 0.092 | si:dkeyp-75h12.5 | -0.049 |
| dut | 0.091 | ckbb | -0.049 |
| pcna | 0.089 | gng3 | -0.047 |
| rrm1 | 0.085 | atp6v0cb | -0.047 |
| anp32b | 0.084 | cadm4 | -0.046 |
| hmgb2b | 0.083 | gpm6ab | -0.046 |
| snrpb | 0.083 | stmn1b | -0.046 |
| ranbp1 | 0.082 | elavl4 | -0.046 |
| banf1 | 0.081 | myt1b | -0.044 |
| chaf1a | 0.081 | atp6v1e1b | -0.044 |
| lig1 | 0.081 | stxbp1a | -0.043 |
| stmn1a | 0.081 | calm1a | -0.043 |
| nutf2l | 0.080 | cotl1 | -0.043 |
| tuba8l4 | 0.080 | actc1b | -0.043 |
| nasp | 0.078 | snap25a | -0.043 |
| mcm7 | 0.078 | stx1b | -0.042 |
| cbx3a | 0.078 | rnasekb | -0.042 |
| sumo3b | 0.078 | pvalb2 | -0.042 |
| ccna2 | 0.078 | acbd7 | -0.041 |
| ran | 0.078 | sncb | -0.041 |
| rpa2 | 0.078 | rbfox1 | -0.041 |
| rrm2 | 0.078 | pvalb1 | -0.041 |
| dek | 0.078 | nsg2 | -0.040 |
| zgc:110540 | 0.077 | elavl3 | -0.040 |
| zgc:56493 | 0.077 | onecut1 | -0.040 |
| hmga1a | 0.077 | rtn1b | -0.040 |
| selenoh | 0.077 | gabarapl2 | -0.040 |
| h2afvb | 0.076 | vamp2 | -0.039 |
| ssrp1a | 0.076 | cplx2 | -0.039 |
| nop58 | 0.076 | ppdpfb | -0.038 |
| snrpf | 0.076 | gapdhs | -0.038 |
| snrpd1 | 0.076 | rtn1a | -0.038 |
| lbr | 0.075 | sncgb | -0.038 |
| mki67 | 0.075 | dpysl3 | -0.038 |
| cks1b | 0.074 | csdc2a | -0.037 |