"zinc finger, SWIM-type containing 8"
ZFIN 
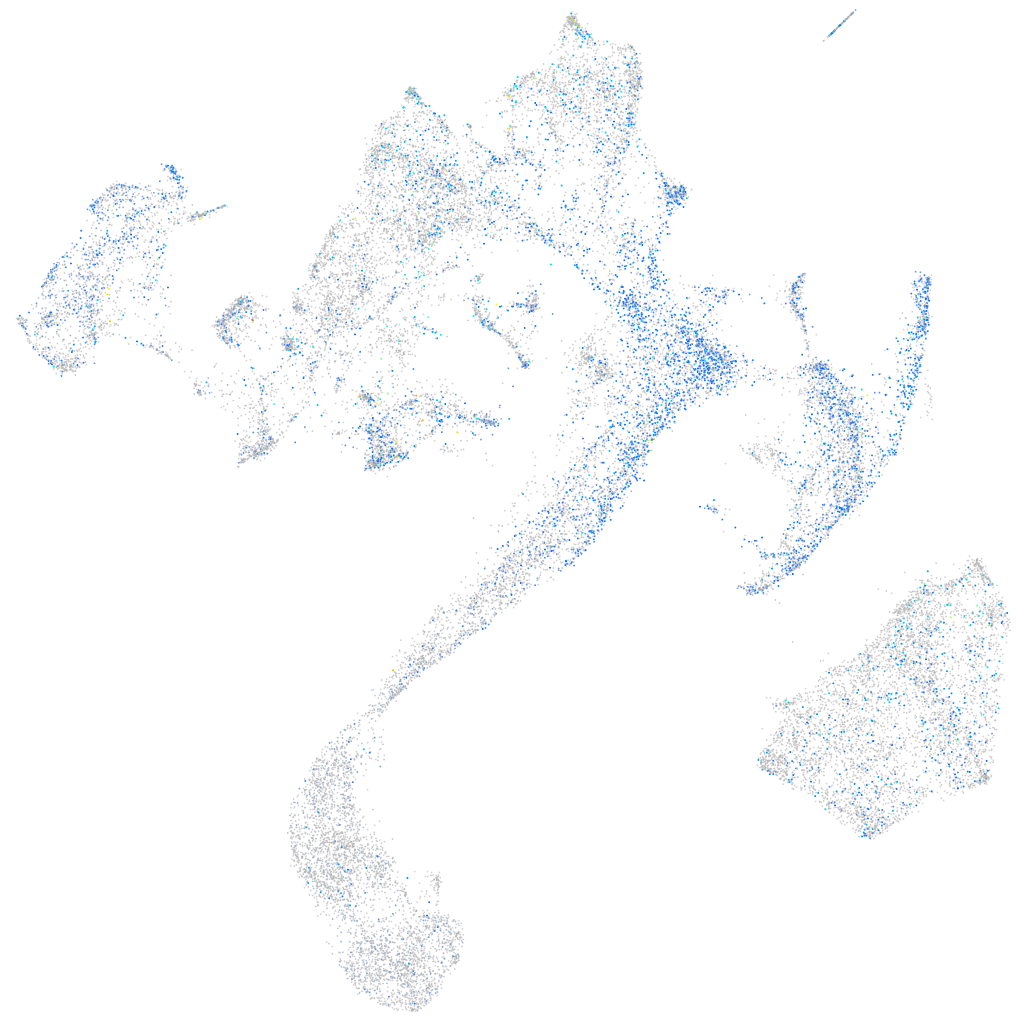
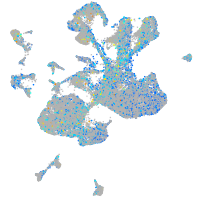
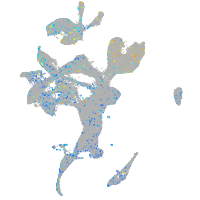
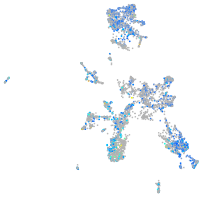
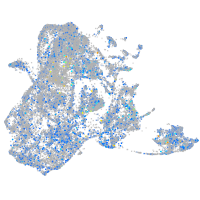
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fn1b | 0.182 | pvalb2 | -0.126 |
| efemp2a | 0.159 | mylz3 | -0.124 |
| XLOC-042229 | 0.152 | pvalb1 | -0.120 |
| selenof | 0.146 | tnni2a.4 | -0.119 |
| si:ch1073-429i10.3.1 | 0.145 | mylpfa | -0.118 |
| cdh2 | 0.144 | mylpfb | -0.118 |
| bmpr1ba | 0.143 | myl1 | -0.117 |
| optc | 0.143 | tnnt3b | -0.116 |
| zbtb18 | 0.140 | myom2a | -0.111 |
| btg2 | 0.139 | myoz1b | -0.111 |
| hnrnpa0l | 0.137 | slc25a4 | -0.106 |
| fbn2b | 0.137 | myhz1.3 | -0.106 |
| qkia | 0.136 | ckmb | -0.104 |
| cpn1 | 0.136 | tnnt3a | -0.104 |
| tspan7 | 0.134 | ckma | -0.104 |
| XLOC-001964 | 0.131 | myhz1.1 | -0.102 |
| XLOC-042222 | 0.130 | si:ch73-367p23.2 | -0.102 |
| sipa1l2 | 0.129 | casq1a | -0.101 |
| plpp1a | 0.126 | CABZ01061524.1 | -0.101 |
| angptl7 | 0.126 | ryr3 | -0.101 |
| mfap2 | 0.126 | hhatla | -0.098 |
| mdka | 0.122 | si:dkey-16p21.8 | -0.097 |
| XLOC-032526 | 0.121 | atp2a1l | -0.095 |
| si:dkey-261h17.1 | 0.121 | si:ch211-255p10.3 | -0.094 |
| rnd3a | 0.121 | tnnc2 | -0.094 |
| h3f3a | 0.121 | atp2a1 | -0.093 |
| emp2 | 0.120 | eef2l2 | -0.092 |
| efnb2a | 0.120 | bhmt | -0.092 |
| net1 | 0.120 | acta1b | -0.091 |
| ubc | 0.120 | tpma | -0.089 |
| ahnak | 0.119 | myhz2 | -0.088 |
| meox1 | 0.119 | myoz1a | -0.086 |
| zgc:56493 | 0.118 | myoz2a | -0.086 |
| naca | 0.117 | prr33 | -0.086 |
| rasgef1ba | 0.117 | ampd1 | -0.085 |